Abstract
Chinese licorice, the dried root of Glycyrrhiza uralensis, is used in traditional Chinese medicine. However, root rot of G. uralensis has gradually become a serious problem in licorice production in China. A 2012 survey of 22 commercial licorice fields revealed a root rot incidence of 36.5% infected plants. Infected plants were wilted with chlorotic foliage and discolored vasculature, and were significantly associated with a scale insect (Porphyrophora ningxiana) that parasitized plant roots (r = 0.9790, P < 0.001).Two previously characterized fungal isolates (FLR and G013) collected from infected licorice plant roots were used to inoculate licorice plant roots, which resulted in symptoms similar to those observed in field plants. Roots of licorice plant inoculated with FLR showed vascular discoloration, whereas those inoculated withG013 resulted in cortical root rot. Using phylogenetic analyses of the ribosomal Intergenic Spacer (IGS), ribosomal DNA and Internal Transcribed Spacers (ITS), translation elongation factor 1-alpha (EF1-a), and RNA polymerase II large subunit (RPB2), isolate FLR was identified as a novel sequence type within the Fusariumoxysporum Species Complex (FOSC), while isolate G013 was identified as a member of F. Solani Species Complex (FSSC) 11. Pathogenicity experiments revealed that isolate FLR can infect and cause disease in melon, soybean and potato; isolate G013 can infect and cause disease in pea, broad bean, soybean and potato. Additional experiments revealed that the incidence of root rot of licorice plants was significantly enhanced when cochineal scales presented in the soil.
Keywords: Glycyrrhiza uralensis; Fusarium oxysporum; Fusariumsolani; Fusarium wilt; Root rot; Porphyrophora ningxiana
Introduction
Glycyrrhiza uralensis Fisch, a leguminous perennial herb, is native to the Middle East and Central Asia including China, Afghanistan, Pakistan, Azerbaijan, Uzbekistan, Turkmenistan, Syria, and Turkey. Licorice, the dried root of G. uralensis, is used in two primary forms: root and extract. Records of licorice plant cultivation date back to the third century [1]. Licorice is a Chinese traditional medicine used for the treatment of various ailments including ulcers, sore throats, arthritis, and allergies [2-4]. It has also been a popular herbal medicine in Europe for centuries [2,5-12]. In addition, licorice is used as a food additive, a raw material in cosmetics and a flavoring agent for tobacco [13-15].
Licorice plant mainly grows in arid desert steppes and was historically harvested from wild resources, mainly limited to northwest and northeast areas of China, before domestic cultivation was established in the 1990s. Licorice plant cultivation has increased dramatically in recent years to fill shortages of wild licorice supplies. For example, licorice was produced on an estimated 125,408 hain 2011 [16]. Since 2002, there have been serious outbreaks of root rot and wilton cultivated G. uralensis plants in Ningxia, China, which severely reduced the yield and quality of licorice. Up to 30% mortality of cultivated licorice plants have occurred in fields and the disease complex is considered to be the most significant threat to commercial production of licorice in this area [17].
Previously, we reported the occurrence of two pathogens associated with Fusarium wilt of licorice plant [17], which were putatively identified as members of the F. oxysporum Species Complex (FOSC) and F. solani Species Complex (FSSC) based on sequence identity of the translation elongation factor 1-alpha (EF-1a) locus [18]. Subsequently, it was observed that 0the death of licorice plant was also associated with Porphyrophora ningxiana Yang (Hemiptera: Margarodidae), a subterranean scale insect commonly referred to as ‘ground pearls’ on account of the nymph stage being enclosed in a pearl-like cyst on the roots of plants. The ‘Cochineal Scale’ (CS), is a sessile plant parasite with one generation each year and four developmental stages: egg, 1st nymph, 2nd nymph (pearl scale), and adult (Figure 1). The CS mainly overwinters as eggs in egg oocysts. In soil, the nymphs can locate and attach to roots to extract sap, and produce the characteristic pearl-like cysts before the growing season commences. In August, adults emerge, mate, and then lay eggs in eggoocystsin soil between 2-6 cm underground [19].
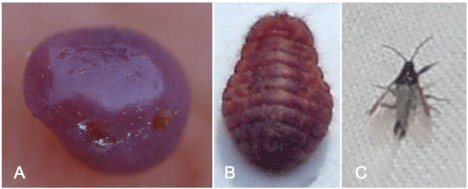
Figure 1: Three morphologies of cochineal scale (Porphyrophora ningxiana Yang). A) 2nd nymph (pearl scale); B) female adult; C) male adult.
The objectives of the current study were to 1) investigate the incidence and density of C Son roots of licorice plants 2) further characterize the Fusarium spp. associated with licorice wilt by performing multilocus phylogenetic analyses 3) assess the effect of licorice plant ground pearl on colonization of Fusarium infections 4) investigate the host range of the Fusarium spp. isolated from licorice plant root.
Materials and Methods
Field incidence of P. ningxiana and Fusarium spp. in cultivated G. uralensis
Symptomatic roots of licorice plants (G. uralensis) were sampled from 22cultivated fields at Ningxia and Inner Mongolia, China in July 2012. Infected plants exhibiting wilt with chlorotic foliage were targeted for destructive sampling. Symptomatic roots were excised for fungal isolation as well as for lab rearing of P. ningxiana (CS) when present. The percent of plants with CS and root discoloration associated with Fusarium infection, mean plant mortality rates, and population densities of insects were recorded.
Collection of P. ningxiana oocysts for laboratory experiments
In 2013, additional P. ningxiana oocysts were collected from cultivated licorice fields. Field soil containing oocysts(c.a. 5 oocysts per 100 g soil) was collected, brought back to laboratory, mixed with cow manure compost and sand (0.1:1:1, v/v/v), and fed and bred in 30-cm-dia. pots containing healthy G. uralensis seedlings. Plants were maintained in a greenhouse with the temperature of 20-30°C and a photoperiod of 10-14 h. Prior to planting, the cow manure compost and sand were autoclaved for 1 hour at 121°C. After c.a. 2 months in the greenhouse, the new oocysts produced in the potted licorice plants were then collected and used for inoculations.
Strain cultivation and inoculum preparation
Isolates from diseased roots were maintained on Potato Dextrose Agar (PDA) amended with 50 ng/ml streptomycin sulfate. Mycelia of FOSC isolate FLR and FSSC isolate G013 were grown in 300ml flasks with 100 ml Potato Dextrose Broth (PDB) at 25°C in darkness for 10days, washed with distilled water twice, and then used for genomic DNA extraction, respectively. Carnation Leaf Agar (CLA) plates were prepared for morphological studies using 15 g agar per 1000 ml distilled water and 4 pieces in 0.5 mm×0.5 mm carnation leaf and were used to induce sporodochium formation and macroconida [20]. The carnation leaves were dried in an oven with temperature of 45-55°C for 2 h and sterilized using Cobalt-60 radiation. Ten-dayold cultures growing on PDA at 25°C in darkness were used to make 5-mm-diameter fungal disks from colony margins using a puncher and transferred to CLA.
DNA manipulations
Genomic DNA was extracted from freeze-dried mycelia using a Hexadecyltrimethylammonium Bromide (CTAB) protocol as described in Kim et al. [21], and purified using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI). The Internal Transcribed Spacer (ITS) regions of the nuclear ribosomal DNA and translation elongation factor 1a (EF-1a) gene were amplified using PCR primers ITS1/ITS4 [22] and EF1-728F/EF1-986R [23], respectively (Table 1). The nuclear ribosomal DNA Intergenic Spacer Region (IGS) was amplified using PCR primers NL11/CNS1 [24] (Table 1). The RNA polymerase II gene (RPB2) was amplified using PCR primers RPB2-5F/RPB2-7cR, and RPB2-7cF/RPB2- 11aR [25] (Table 1). PCR amplifications were done with Platinum Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA) in an Applied Biosystems 9700 thermo cycler (CA, USA) using the following program: 1 cycle of 2 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C; followed by 1 cycle of 10 min at 72oC and a 4oC forever. All PCR products were separated in a 1.5% agarose gel, stained with ethidium bromide, and photographed over a UV transilluminator. PCR products were purified using the Agarose Gel Extraction Kit (Roche Inc., Germany), cloned into pDM18-T vector (TaKaRa, Japan) and transformed into Escherichia coli JM109 using an electroporation method [26]. After cloning, 10 positive colonies from each transformation were randomly selected, and each colony was recovered in a LB liquid medium supplemented with 100 μg.ml-1 ampicillin following overnight incubation at 37°C. The recombinant plasmid was extracted using Plasmid Miniprep Kit (Bioniga, USA). The confirmation of positive colonies was confirmed through restriction digestion with KpnI and Hind III. The plasmid was sequenced by Life technologies (Thermo Fisher Scientific, USA).
Primer name
Primer sequence5'-3'
Loci amplified
Annealing temperature (oC)
Primer sequence origin
ITS1
TCCGTAGGTGAACCTGCGG
ITS
55
White, 1990
ITS4
TCCTCCGCTTATTGATATGC
EF1-728F
CATCGAGAAGTTCGAGAAGG
EF-1a
55
Carbone et al., 1999
EF1-968R
TACTTGAAGGAACCCTTACC
NL11
CTGAACGCCTCTAAGTCAG
IGS
55
O’Donnell et al., 2009
CNS1
GAGACAAGCATATGACTACTG
RPB2-5F
GA(T/C)GA(T/C)(A/C)G(T/A)GATCA(T/C)TT(T/C)GG
RPB2
55
Liu et al., 1999
RPB2-7cR
CCCAT(A/G)GCTTG(T/C)TT(A/G)CCCAT
RPB2-7cF
ATGGG(T/C)AA(A/G)CAAGC(T/C)ATGGG
RPB2
55
Liu et al., 1999
RPB2-11aR
GC(A/G)TGGATCTTRTC(A/G)TC(C/G)ACC
M13F(-47)
CGCCAGGGTTTTCCCAGTCACGAC
-
55
pMD18-T, TaKaRa
M13R(-48)
AGCGGATAACAATTTCACACAGGA
FLR.W1F
TGGCGGATCTGACACTGTCG
-
51
Primer walking sequence
FLR.W1R
CACGCCAGAACTGCTTCGTG
G013.W1F
CCACCAATGCCGCCATTCTT
-
49
Primer walking sequence
G013.W1R
GCATCCTCAAGGCACCAACA
Table 1: Primers used in this study.
Phylogenetic analysis
Isolates FLR and G013 were previously identified as “F. oxysporum” and “F. solani”, respectively, based on EF-1a sequences. In this study, additional loci were sequenced and analyzed using existing Fusarium data sets for phylogenetic species recognition based on genealogical concordance of multilocus DNA sequence data [24,27]. This approach has previously been used to identify environmental isolates of Fusarium spp. to species and sequence type [28]. Two nucleotide sequence data sets, one for FOSC and one for FSSC were constructed from data in NCBI GenBank; additional sequence data was kindly provided by Dr Kerry O’Donnell (USDAARS). Sequence data for EF-1a and IGS from isolate FLR were aligned with the data set of known FOSC diversity using Molecular Evolutionary Genetics Analysis (MEGA) 6.06 [29]. Similarly, sequence data for EF-1a, ITS and RPB2 were aligned with the data set of known FSSC diversity. Maximum Parsimony (MP) and Maximum Likelihood (ML) analyses were performed on both data sets; insertion deletion polymorphisms were not coded because MEGA does not allow for numerical characters. MP phylogenies were constructed using a sub tree-pruning-regrafting search method with 10 initial trees, a search level of 1, and 100 maximum trees to retain; 100 bootstrap replications were performed. ML phylogenies were constructed using a Tamura-Nei substitution model, uniform rates among sites, a heuristic method of nearest-neighbor-interchange, an initial neighbor joining tree, and a very strong branch swap filter; 100 bootstrap replications were performed. After the aforementioned phylogenetic analyses, sequence data for isolates FLR and G013 were visually compared to the most similar FOSC and FSSC sequence types in respective phylogenetic trees, in order to verify that they contained unique characters that differentiated them from other sequence types.
Effect of root feeding by cochineal scale on Fusarium infection
To determine the relationships between licorice plants infected by Fusarium spp. And CS colonization, the following four treatments were initiated: for treatment I: licorice seedlings were planted in pots of 30-cm-diameter (4 seedlings each) containing cow dung compost and sand (1:1, v/v) as a non-inoculated control; for treatment II: five petri dishes (9 cm in diameter) of 3-days PDA cultures of FLR and G013 were cut into small pieces using a sterilized scalpel and mixed into cow dung compost and sand of each pot. The oocysts of CS (ca. 20) were buried near the root of licorice seedlings at an approximate depth of 2-10 cm; for treatment III: the same as in treatment II except without oocysts of CS; for treatment IV: same as treatment II except that oocysts of CS were excluded and roots were wounded with a sterilized needle 2-10 cm below the soil surface. Each treatment contained two replicates for a total of eight pots (ca. 30 seedlings) across all treatments. All seedlings were kept in a greenhouse with the temperature of 20-30°C and a photoperiod of 10-14 h.
Host range of Fusarium isolates
Inoculations with FOSC isolate FLR and FSSC isolate G013 were conducted in the greenhouse on 1-month-old potted G. uralensis seedlings (12 plants per treatment) to confirm pathogenicity. These served as a positive control for the other species tested. One root per seedling was wounded about 2-10 cm below the soil surface using a sterilized needle. Fungal isolates were started on PDA and incubated in darkness at 22°C for 3 days. Five 5-mm-diameter fungal disks from colony margins were taken and firmly placed on the wounded root of each plant and fixed with aluminum foil. Seedlings treated with sterile PDA disks were used as controls. Root disease was assessed at two months after inoculation. To complete Koch’s postulates, the inoculated isolates were re-isolated and confirmed as the pathogen inoculated morphologically.
To determine whether Fusarium spp. isolates from licorice plant root caused disease on other members of the Fabaceae, other plants including alfalfa (Medicago sativa L.), kidney bean (Phaseolus vulgaris L.), pea (Pisum sativum L.), broad bean (Viciafaba L.), cowpea (Vigna unguiculata (L.) Walp.), soybean (Glycine max (L.) Merr.). Melon (Cucumismelo L.), cucumber (Cucumis sativus L.), eggplant (Solanum melongena L.), potato (Solanum tuberosum L.), pepper (Capsicum frutescens L.), tomato (Solanum lycopersicum L.), wheat (Triticum aestivum L.), corn (Zea mays L.) and cotton (Gossypium hirsutum L.) were used in the host range experiments. Commercial seeds were purchased from Xi’an Gude Seed Company, China. Healthy seedlings of G. uralensis supplied by Gancao Experimental Station of Yanchi County, China were included as positive controls. The extent of root and crown discoloration in inoculated and control plants was rated on a 0-to-5 scale, where 0 = no vascular discoloration; 1 = 1 to 25% of the vascular tissue exhibiting discoloration; 2 = 26 to 50%, 3 = 51 to 75%, and 4 = 76 to 100% discoloration; and 5 = 100% discoloration.
The disease scale values of each plant in each treatment were scored and averaged.
Data analysis
Pearson correlation analysis was conducted to determine the relationship between diseased licorice plants and P. ningxianainhabited roots. All data analysis was conducted using the software R (version 2.11.1). The sequences of Internal Transcribed Spacer (ITS), intergenic spacer region (IGS), translation elongation factor 1a (EF-1a), and the RNA polymerase II gene (RPB2) gene were used to construct the phylogenetic tree using Molecular Evolutionary Genetics Analysis (MEGA) 6.06.
Results
Field incidence of Fusarium spp. and P. ningxiana in cultivated G. uralensis
Typical symptoms of Fusarium root rot of G. uralensis were observed in Yanchi county, Ningxia, China. Initial symptoms included wilting and chlorosis of foliage as well as obvious root rot as a severe brown discoloration of vascular tissue (Figure 2A,B). White mycelia were visible on diseased root tissue and roots were eventually decomposed.
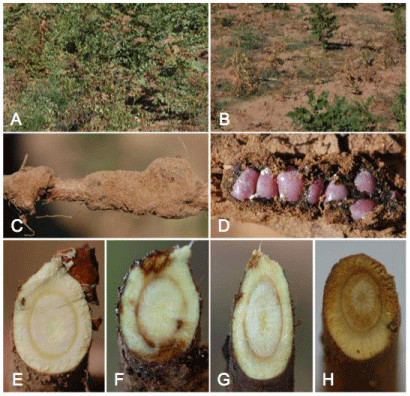
Figure 2: Symptoms of Glycyrrhiza uralensis root rot caused by Fusarium spp. A) healthy plants in a field; B) diseased plants in a field; C) root colonized by cochineal
scales; D) cochineal scales; E) cross-section of a healthy root; F) cross-section of a diseased plant colonized by cochineal scales; G) vascular discoloration of root
produced by isolate FLR one months after inoculation; H) cortex tissue was destroyed, and eventually all root tissue by isolate G013 one months after inoculation.
The association of root discoloration with root rot symptoms varied from 0 to 92% (mean = 36.5%, n=479) (Table 2). The cooccurrence of diseased roots and P. ningxiana (Figure 2C,D) varied from 0.0 to 84.0% (mean = 29.7%, n = 479) (Table 2). The association of plant mortality with diseased roots varied from 0 to 50% across all sampled locations (mean = 11%, n = 479) (Table 2). Diseased licorice plants (root discoloration rate) had a significant relationship with P. ningxiana-inhabited roots (plant rate with insect) (r=0.9790, P<0.001, n = 11).
Site
Latitude and longitude
Altitude (m)
Number of plant
Plant with insect rate (%)
Root discoloration rate (%)
Plant death rate (%)
Population density of insect
Growth type
Zuojiwan, Yanchi, Ningxia
N37°59'317”, E107°21'962”
1334
50
84
92
50
5.8
C
Wanglejin, Yanchi, Ningxia
N37°53'509”, E106°59'845”
1416
46
0
0
0
0
W
Gaoshawo, Yanchi, Ningxia
N37°59'270”, E107°02'461”
1413
40
12.5
37.5
5
2.2
W
Liubazhuang, Yanchi, Ningxia
N37°50'498”, E107°18'120”
1433
16
18.8
25
0
4.1
W
Habahu, Yanchi, Ningxia
N37°42'580”, E107°03'407”
1453
50
0
0
0
0
W
Sunjialoua, Yanchi, Ningxia
N37°51'841”, E107°03'078”
1476
30
73.3
83.3
40
16.1
C
Sunjialoub, Yanchi, Ningxia
N37°52'843”, E107°03'008”
1476
48
27.1
31.3
0
9
W
Chengxitan, Yanchi, Ningxia
N37°47'818”, E107°17'327”
1429
66
0
0
0
0
C
Shabianzi, Yanchi, Ningxia
N37°53'549”, E107°31'97”
1322
50
84
90
8
41.7
N
Aolezhaoqi, Inner Mongolia
N38°10'027”, E107°27'702”
1319
33
27.3
42.4
18.2
22.3
W
Sanduandi, Inner Mongolia
N38°08'758”, E107°25'697”
1314
50
0
0
0
0
W
Average
-
-
-
29.7
36.5
11
9.2
-
C: artificially cultivated Glycyrrhiza uralensis plants; W: wild Glycyrrhiza uralensis plants; N: Glycyrrhiza uralensis plants in nursery.
Table 2: Occurrence of Porphyrophora sophorae and Fusarium spp. on Glycyrrhiza uralensis plants in Ningxia and Inner Mongolia, China.
Pathogen isolation and identification
Isolates from diseased roots were grown on PDA amended with streptomycin sulfate. Isolates (n = 80) previously recovered from symptomatic roots (n= 105) [17] were single-spored to obtain pure cultures. The most frequently isolated fungal morphotypes were identified putatively as F. oxysporum (30.8%) and F. solani (61.5%) based on morphological characteristics including macrocondia and monophialides [30]. Roots of licorice plant inoculated with FLR showed vascular discoloration (Figure 2E,F,G). Roots inoculated with G013 showed cortical root rot (Figure 2H). Fusaria with typical morphology were re-isolated from symptomatic roots. Control plants inoculated with sterile PDA disks remained asymptomatic two months post inoculation and no pathogen was isolated.
Morphological features of G013 were as follows: top of colonies on PDA were white to light yellow-green, with purple-red pigment in the center (Figure 3A); reverse of colonies were yellow to light yellow (Figure 3B). Fusiform macroconidia of G013 were 20.0-50.0×4.5- 7.5μm, rounded at both ends, 2-5 septa and with obvious podocytes (Figure 3C). Kidney-shaped microconidia of G013 were7.5- 16.0×2.5-5.3μm, formed on long, septate and occasionally branched conidiophores (Figure 3D,E). Colonies of FLR on PDA were white, with a purple pigment in center (Figure 4A); reverse of colonies were yellow to light yellow (Figure 4B). Conidiophores were 7.5-22.5×2.5- 5.0μm, solitary, unbranched, and slightly smaller near the point of sporulation (Figure 4C). Microconidia of FLR were 5.0-15.0×2.5- 7.5μm, ovoid, aseptate or occasionally single septate, born on lateral, bottle-shaped conidiophores (Figure 4D). Macroconidia of FLR were 15.0-32.5×5.0-10.0μm, sickle-shaped, and usually 3 to 5 septa (Figure 4E). Chlamydospores were spherical, borne on or between the apical short branches, either alone or in chains (Figure 4F).
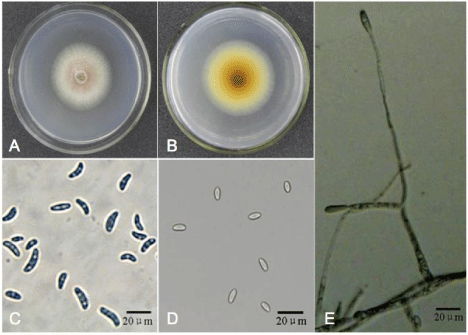
Figure 3: Morphology of isolate G013. A) the front of the colony after being cultivated on Potato Dextrose Agar (PDA) plate in darkness for 2 days; B) the back of
the colony after being cultivated on PDA plate in darkness for 2 days; C) microconidia; D) macroconidia; E) conidiophores.
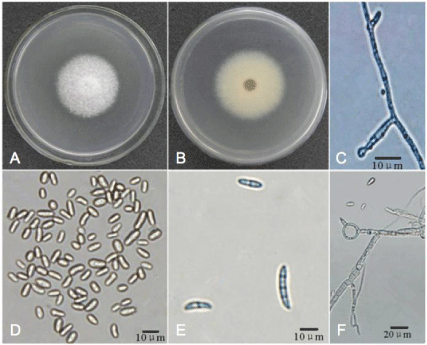
Figure 4: Morphology of isolate FLR. A) the front of the colony after being cultivated on Potato Dextrose Agar (PDA) plate in darkness for 2 days; B) the back of
the colony after being cultivated on PDA plate in darkness for 2 days; C) conidiophores; D) microconidia; E) macroconidia; F) chlamydospore.
Taxonomic statue
Inoculated plant
DS
Latin name
Cultivar
Common name
G013
FLR
Leguminosae
Medicago sativa L.
Muxuwang
Alfalfa
0
0
Phaseolus vulgaris L.
Fengshou 1
Kidneybean
0
0
Pisum sativum L.
Wangnong 604
Pea
1.1
0
Vicia faba L.
Qinghai 3
Broad bean
0.8
0
Vigna unguiculata (L.) Walp.
Zaoyu 80
Cowpea
0
0
Cucurbitaceae
Cucumismelo L.
Zaomi 1
Melon
0
2.8
Cucumissativus L.
Jinyou 1
0
0
Papilionaceae
Glycine max (L.) Merr.
Huajiang 4
0.9
0.8
Solanaceae
Solanummelongena L.
Liuyeqie
Eggplant
0
0
Solanum tuberosum L.
Zhuangshu 4
0.8
3.3
Capsicum frutescens L.
8819
Pepper
0
0
Solanum lycopersicum L.
Zhongza 109
Tomato
0
0
Gramineae
Triticumaestivum L.
Xiaoyan 22
Wheat
0
0
Zea mays L.
Shandan 609
Corn
0
0
Malvaceae
Gossypium spp.
Jimian 11
Cotton
0
0
DS = Disease severity was assessed based on the extent of root and crown discoloration on a 0-to-5 scale, where 0 = no vascular discoloration; 1 = 1 to 25% of the vascular tissue exhibiting discoloration; 2 = 26 to 50%, 3 = 51 to 75%, and 4 = 76 to 100% discoloration; and 5 = 100% discoloration.
Table 3: The host range of the pathogenic isolate G013 and FLR.
Phylogenetic species identification based on genealogical concordance of multilocus DNA sequence data clearly identified isolate G013 as a member of FSSC 11, also known as F. solanif. sp. Pisi and mating population VI [31]. Several unique Single Nucleotide Polymorphisms (SNPs) and insertion deletion polymorphisms (indels) within the multilocus data set were unique, revealing that G013was a novel sequence type within FSSC 11. The FOSC isolate FLR appeared as a distinct sequence type within FOSC; while this isolate shared an identical EF-1a allele and nearly identical IGS allele with NRRL 32328 (ST 197), that contained a unique 79-bp insertion polymorphism within the IGS (Figure 5).
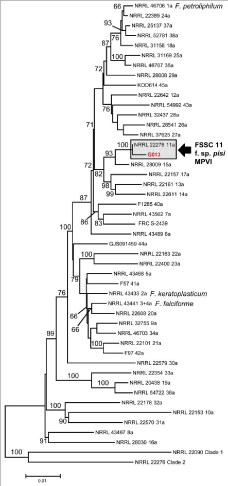
Figure 5: Phylogenetic tree of the Fusariumsolani Species Complex (FSSC)
constructed using Maximum Likelihood. Isolate G013 is conspecific with
FSSC 11, F. solani f. sp. pisi, (Mating population VI and MPVI). Alignment
included 3441 characters.
Effect of cochineal scale colonization on Fusarium infection
Percent disease incidence in G. uralensis seedlings in treatment I, II, III, and IV was 0.0, 67.7, 10.0, and 86.5, respectively. Plants wounded and inoculated with FLR and G013 (treatment IV) showed the highest disease incidence, which has no significant difference (P>0.01) with treatment II; however, both treatments II and IV were significantly different (P<0.01) from treatment III and the control (treatment I) (Figure 6), suggesting that the incidence of root rot of licorice seedlings was greatly enhanced by the co-occurrence of CS in infested soil.
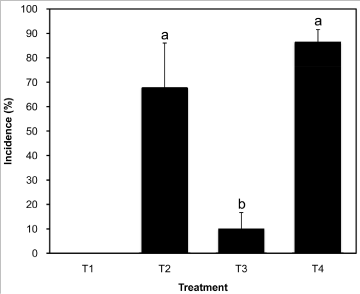
Figure 6: Incidence of Glycyrrhiza uralensis caused by Fusarium oxysporum
and F. solani enhanced by cochineal scales. T1, healthy G. uralensis plants
were planted into plots with sterilized sand-soil; T2, healthy G. uralensis
plants were planted into plots with sterilized sand-soil, cochineal scales, and
FLR and G013 (vol: vol, 1:1); T3, healthy G. uralensis plants were planted
into plots with sterilized sand-soil, and FLR and G013 (vol: vol, 1:1); T4,
healthy G. uralensis plants wounded by sterilized needle on the root was
planted in plots with sterilized sand-soil, and FLR and G013(vol: vol, 1:1).
Host range of Fusarium isolates
Pathogenicity tests with the two representative Fusarium isolates were conducted in a greenhouse on 1-month-old potted seedlings of Chinese licorice, alfalfa, kidney bean, pea, soybean, broad bean, cowpea, melon, cucumber, eggplant, potato, pepper, tomato, wheat, corn, and cotton (12 plants per crops). FSSC strain G013 infected and caused disease in pea, broad bean, soybean, and potato with disease severity of 1.1, 0.8, 0.9 and 0.8 respectively, while FOSC strain FLR infected and caused disease in melon, soybean, and potato with disease severity of 2.8, 0.8 and 3.3 respectively (Table 3).
Discussion
Insects can play a key role in the emergence of landscapewide diseases [32]. Fusarium spp. Has been implicated in several landscape-wide diseases involving insects [33-35]. Insects can play a significant role in wounding plant tissue, providing infection courts for soil-borne Fusarium spp. In the soil [36,37].
The F. solani species complex comprises over 50 phylogenetic species, the majority of which have not been formally described [31]. The complex comprises many well-known plant pathogens that cause diseases on a wide range of plants [38]. Members of the FSSC are cosmopolitan in soils and cause tuber, root and stem rots of plants worldwide [39]. In the current study, one of the Fusaria isolated from Chinese licorice plants was conspecific with FSSC 11, also known as F. solani f. sp. Pisi or F. solani mating population VI. Members of FSSC 11 are known to deactivate phytoalexins, such as pisatin in pea [40,41]. Medicarpin 7, an isoflavone from licorice, may serve a similar role to pisatin, and therefore, we speculate that FSSC 11 may utilize a similar deactivation mechanism in licorice plant [42]. Further, the connection between F. solani f. sp. pisi, which is a known as pea pathogen, and G. uralensis, may indicate the presence of conserved pathogenic mechanisms, given that G. uralensis is also a leguminous plant in the Fabaceae.
F. oxysporum complex is a group of cosmopolitan soilborne Fusaria that act as both saprophytes and plant pathogens [43]. The FOSC can cause severe vascular wilts and yield losses in various crops including banana, pepper, potato, strawberry, tomato and watermelon [44]. While members of the FSSC generally cause plant disease through necrosis of roots and other subterraneous tissue, the FOSC hyphae can directly penetrate the root epidermal walls, then advance intercellularly through the root cortex and into xylem vessels [36,45]. Fusarium wilt symptoms are compounded by water stress and are the result of occlusion of vessels by fungal mycelium accumulation, which include the production of gels, tyloses and proliferation of parenchyma cells [46]. The disease symptoms observed in licorice plant were similar: leaf chlorosis and unequal wilting of the aerial parts in the early stages, and eventual collapse and death of the plant.
Wounds in licorice plant roots such as those caused by CS may provide an infection court for the pathogen [47]. This study provides the first evidence that ground pearl feeding activity on plant tissue in the presence of fungal inoculum can result in higher incidence of Fusarium wilt. Fusarium wiltis an economically significant problem affecting licorice production of Ningxia and Inner Mongolia, China that first occurred in 2002. The area affected by Fusarium wilt has expanded in recent years. We performed preliminary evaluations of carbendazim, daconil, metalaxylmancozeb, triophanate-methyl, and fludioxonilonFRL and G013 and thiamethoxam, imidacloprid, chlorpyrifos, tianjielongon CS. The combination of thiamethoxam and metalaxylmancozebappears to have potential in controlling Fusarium wilt associated with CS (data not shown). Rotation with high sulfur glycoside-containing rape varieties is another option [48]. Soil fumigation using methyl bromide (MeBr) was mainly used in controlling soilborne diseases in the past several decades [49], but has been identified as an ozone depleting substance and is currently being phased out of use. Once alternatives to MeBr are approved, additional tools to manages oilborne diseases will be available [50,51].
Conclusion
The root rot of Chinese licorice plant was caused by F. solani Species Complex (FSSC) 11 and Fusarium oxysporum Species Complex (FOSC). The disease was significantly enhanced when cochineal scales presented in the soil. The strain FLR can infect and cause disease in melon, soybean and potato; isolate G013 can infect and cause disease in pea, broad bean, soybean and potato. The efficiency control measures including fungicides, such as metalaxyl mancozeb, daconil, thiophanate-methyl, et al., should be established as soon as possible.
Acknowledgment
The study was funded by Ningxia-Shaanxi Science and Technology Cooperative grand and by the Key Laboratory of Urban Agriculture (North) Ministry of Agriculture (KFK2015001).
References
- Olukoga A, Donaldson D. Historical perspectives on health. The history of liquorice: the plant, its extract, cultivation, commercialization and etymology. J Roy Soc Health. 1998; 118: 300-304.
- Asl MN, Hosseinzadeh H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother Res. 2008; 22: 709-724.
- Gao X, Wang W, Wei S, Li W. Review of pharmacological effects of Glycyrrhiza radix and its bioactive compounds. China J Chinese Materia Medica. 2009; 34: 2695-2700.
- Chinese Pharmacopoeia Commission. People's Republic of China Pharmacopoeia. Beijing: China Medical Science Press. 2010.
- Chien CF, Wu YT, Tsai TH. Biological analysis of herbal medicines used for the treatment of liver diseases. Biomed Chromatogr. 2011; 25: 21-38.
- Hasani-Ranijbar S, Nayebi N, Moradi L, Mehri A, Larijani B, Abdollahi M. The efficacy and safety of herbal medicines used in the treatment of hyperlipidemia: a systematic review. Curr Pharm Design. 2010; 16: 2935-2947.
- Gazzani G, Daglia M, Papetti A. Food components with anticaries activity. Curr Opin Biotechnol. 2012; 23: 153-159.
- Messier C, Epifano F, Genovese S, Grenier D. Licorice and its potential beneficial effects in common oro-dental diseases. Oral Dis. 2012; 18: 32-39.
- Sato H, Goto W, Yamamura J, Kurokawa M, Kageyama S, Takahara T, et al. Therapeutic basis of glycyrrhizin on chronic hepatitis B. Antiviral Res.1996; 30:171-177.
- Wu TY, Khor TO, Saw CL, Loh SC, Chen AI, Lim SS, et al. Anti-inflammatory/Anti-oxidative stress activities and differential regulation of Nrf2-mediated genes by non-polar fractions of tea Chrysanthemum zawadskii and Licorice Glycyrrhiza uralensis. AAPS J. 2011; 13: 1-13.
- van Rossum TG, Vulto AG, de Man RA, Brouwer JT, Schalm SW. Glycyrrhizin as a potential treatment for chronic hepatitis C. Aliment Pharmacol Ther. 1998; 12:199-205.
- Yasui S, Fujiwara K, Tawada A, Fukuda Y, Nakano M, Yokosuka O. Efficacy of intravenous glycyrrhizin in the early stage of acute onset autoimmune hepatitis. Dig Dis Sci. 2011; 56: 3638-3647.
- Carmines EL, Lemus R, Gaworski CL. Toxicologic evaluation of licorice extract as a cigarette ingredient. Food Chem Toxicol. 2005; 43: 1303-1322.
- Cook MK. Makes natural sweeteners more versatile? Food Engineering. 1973; 45: 145-146.
- Tilley NM. The bright tobacco industry, 1860-1929. Chapel Hill: University of North Carolina Press. 1948.
- Chen KZ, Hu S, Chen R. Licorice industry in China: implications for licorice producers in Uzbekistan. International Food Policy Research Institute Beijing, China. 2014.
- Cao XM, Cai J, Li SB, Zhang H, Lu ZQ, Hu XP. Fusarium solani and Fusarium oxysporum associated with root rot of Glycyrrhiza uralensis. Plant Dis. 2013; 97: 1514.
- Geiser DM, Jiménez-Gasco MDM, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, et al. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur J Plant Pathol. 2004; 110: 473-479.
- Yang C, Gao L, Zhang Z. Study on the occurrence rules and the technology of integrated control of Porphyrophora ningxiana. World SciTech. 2006; 8: 128-135.
- Fisher NL, Burgess LW, Toussoun TA, Nelson PE. Carnation leaves as a substrate and for preserving cultures of Fusarium species. Phytopathology. 1982; 8: 1041-1049.
- Kim DH, Martyn RD, Magill CW. Restriction fragment length polymorphism groups and physical map of mitochondrial DNA from Fusarium oxysporum f. sp. niveum. Phytopathology. 1992; 82: 346-353.
- White TJ, Bruns SL, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications, Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Academic Press, Inc, New York. 1990; 315-322.
- Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999; 91: 553-556.
- O’Donnell K, Gueidan C, Sink S, Johnston PR, Crous PW, Glenn A, et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet Biol. 2009; 46: 936-948.
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationship among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999; 16: 1799-1808.
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988; 16: 6127-6145.
- O’Donnell K, Sutton DA, Fothergill A, McCarthy D, Rinaldi MG, Brandt ME, et al. Molecular phylogenetic diversity, multilocus haplotype nomenclature, and in vitro antifungal resistance within the Fusarium solani species complex. J Clin Microbiol. 2008; 46: 2477-2490.
- Short DPG, O’Donnell K, Zhang N, Juba JH, Geiser DM. Widespread occurrence of diverse human pathogenic types of the fungus Fusarium detected in plumbing drains. J Clin Microbiol. 2011; 49: 4264-4272.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725-2729.
- Booth C. The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, UK. 1971.
- O’Donnell K. Molecular phylogeny of the Nectria haematococca-Fusarium solani species complex. Mycologia. 2000; 92: 919-938.
- Dunn RR, Hulcr J. The sudden emergence of pathogenicity in insect-fungus symbioses threatens naive forest ecosystems. Proc Roy Soc B. 2011; 278: 2866-2873.
- Kasson MT, O’Donnell K, Rooney AP, Sink S, Ploetz RC, Ploetz JN, et al. An inordinate fondness for Fusarium: Phylogenetic diversity of fusaria cultivated by ambrosia beetles in the genus Euwallacea on avocado and other plant hosts. Fungal Genet Biol.2013; 56:147-157.
- Sakamoto JM, Gordon TR, Storer AJ, Wood DL. The role of Pityophthorus spp. as vectors of pitch canker affecting Pinusradiata. Can Entomol. 2007; 139: 864-871.
- O’Donnell K, Sink S, Libeskind-Hadas R, Hulcr J, Kasson MT, Ploetz RC, et al. Discordant phylogenies suggest repeated host shifts in the Fusarium-Euwallacea ambrosia beetle mutualism. Fungal Genet Biol. 2014; 99: 277-290.
- Mendgen K, Hahn M, Deising H. Morphogenesis and mechanisms of penetration by plant paghogenic fungi. Annu Rev Phytopathol. 1996; 34: 364-386.
- Scarlett K, Tesoriero L, Daniel R, Guest D. Sciarid and shore flies as aerial vectors of Fusarium oxysporum f. sp. cucumerinum in greenhouse cucumbers. J Appl Entomol. 2014; 138: 368-377.
- Kolattukudy PE, Gamble DL. Nectria haematococca: pathogenesis and host specificity in plant diseases. In: Pathogenesis and Host Specificity in Plant Pathogenic Fungi and Nematodes, Eukaryotes. Kohmoto K, Singh US, Singh RP, editors. Pergamon Press, Oxford, UK. 1995: 2: 83-102.
- Desjardins AE. Fusarium mycotoxins: chemistry, genetics and biology. The American Phytopathological Society, APS Press, St. Paul, MN. 2006.
- Ciuffetti LM, VanEtten HD. Virulence of a pisatin demethylase-deficient Nectria haematococca MPVI isolate is increased by transformation with a pisatin demethylase gene. Mol Plant-Microbe Interact. 1996; 9: 787-792.
- Wasmann CC, VanEtten HD. Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol Plant-Microbe Interact. 1996; 9: 793-803.
- Akashi T, VanEtten HD, Sawada Y, Wasmann CC, Uchiyama H, Ayabe S. Catalytic specificity of pea O-methyltransferases suggests gene duplication for (+)-pisatin biosynthesis. Phytochemistry. 2006; 67: 2525-2530.
- Zhang N, O’Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye AA, et al. Members of the Fusariumsolani species complex that cause infections in both humans and plants are common in the environment. J Clin Microbiol. 2006; 44: 2186-2190.
- Nelson PE. Life cycle and epidemiology of Fusarium oxysporum. In: Fungal Wilt Diseases of Plants. Mace ME, Bell AA, Beckman CH, editors. Academic Press, New York, NY. 1981.
- Bishop CD, Cooper RM. An untrastructural study of vascular colonization in three vascular wilt diseases I. Colonization of susceptible cultivars. Physiol Mol Plant Pathol. 1982; 23: 323-343.
- Beckman CH. The nature of wilt diseases of plants. The American Phytopathological Society, APS Press, St. Paul, MN. 1987.
- Dwinell LD, Fraederich SW, Adams D. Diseases of pines caused by the pitch canker fungus. In: Fusarium. Paul Nelson Memorial Symposium. Summerell BA, Leslie JF, Backhause D, Bryden WL, Burgess LW, editors. APS Press, St. Paul, MN. 2001: 225.
- Brown PD, Morra MJ. Control of soilborne plant pests using glucosinolate-containing plants. Adv Agron. 1997; 61: 167-231.
- Martin FN. Development of alternative strategies for management of soilborne pathogens currently controlled with methyl bromide. Annu Rev Phytopathol. 2003; 41: 325-350.
- Duniway JM. Status of chemical alternatives to methyl bromide for pre-plant fumigation of soil. Phytopathology. 2002; 92: 1337-1343.
- Meadows R. News overview: Researchers develop alternatives to methyl bromide fumigation. California Agriculture. 2013; 67: 125-127.
