Abstract
Most of the local strawberry (Fragaria x ananassa Duchenese) cultivars of Turkey are not phylogenetically classified. This present study, which aimed discriminating the genetical relationships between the Ottoman strawberry cultivar and other early-period strawberry cultivars planted in Istanbul and Karadeniz Ereğli regions of Turkey, is the first on this field. Seven strawberry cultivars were analyzed by using RAPD. 10 random primers carried out DNA fingerprinting analyses of these cultivars. The average polymorphism rate was determined as 87.1%. The most polymorphic primer was OPH01 which produced 13 bands. The cluster dendrogram presented that the similarity coefficients were between range of 0.03 and 0.73. Ottoman cultivar showed higher similarity with Tüylü more than Kara cultivar which is reported as the mother cultivar. RAPD method was sufficient to assess the phylogenetic relationship between Ottoman cultivar and other early-period strawberry cultivars. The data of this study brought forward the necessity of further genetic analysis to prove the phylogenetic relationship among Tüylü and Ottoman cultivars.
Keywords: Ottoman strawberry cultivar; Early-period strawberry; Genetic diversity; RAPD
Introduction
Strawberry is a hybrid species of Fragaria genus (Fragaria x ananassa Duchesne). It is widely cultivated worldwide for its characteristic aroma, color, juicy texture and sweetness. The strawberry fruit has been used in medicine during ancient Rome. Its cultivation became more common after 16th century. New species were gradually dispersed to the world at the end of 18th century. It is consumed fresh or frozen and used in processed food products such as cereal bars, ice cream, flavored milk and yogurt.
The important characteristics of strawberry are the fragrance and the flavor. Planting conditions and variety based changes effect the taste of strawberry via altering the sugar, acids and volitant compounds of the strawberry. Turkey is among the largest strawberry producers and consumers following USA, Spain, Italy and Japan. Although it is greatly consumed all over the world. In these countries, the breeding and cultivation programs focus on yield, disease resistance and enhancement of shelf-life [1]. In Turkey, studies on strawberries mostly focus on yield, aroma and taste.
All the cultivars have unique morphological features. Ottoman strawberry has more specificity since it takes place in endangered species nowadays.
In the late 1700s, strawberry was first brought from Europe to Turkey. Cultivation of this newcomer was planted around Arnavutköy region of Istanbul and therefore named as Arnavutköy cultivar. In Karadeniz Ereğli, there are some early period strawberry cultivars but there is no documentary of these cultivars related to their origin (hybrids or wild types). Ottoman strawberry which has specific taste and smell has been rumored to be a hybrid of “Arnavutköy” and “Kara” cultivars. Today more than 500 families farming Ottoman strawberry in Karadeniz Ereğli region. The planting process needs more attention in this region. (ARAYA BIR GEÇIS CÜMLESI OLSA) the fruits are harvested till the half of the June.
There is no scientific data related to the genetic diversity of “Ottoman” and other local strawberry cultivars. They are not produced commercial scale and the only data mentioning the “Arnavutköy” cultivar was collected just to improve some new cultivars in Yalova, Turkey. The phylogenetic relationship of Ottoman cultivar has not been characterized. This is the first study to assess the genetic divergence of Ottoman strawberry with other early-period strawberry varieties.
New breeding programs have been developed for strawberry to improve new cultivars adapted to different climates and having different characteristics. At this point, identification of the germplasms by molecular methods is essential. Patented strawberry varieties which have economical importance have been evaluated for their genetic diversity and the molecular patterns of each cultivar [1,2]. Several techniques based on DNA analysis are available for genetic diversity studies. Randomly Amplified Polymorhic DNA (RAPD) is an efficient molecular marker technique for genotyping and phylogenetic analysis. It is widely preferred since being rapid, economic and no requirement to previous information of the genome to evaluate different organisms [3-5].
RAPD (Random Amplification of Polymorphic DNA) is a widely used method for genetic diversity studies in strawberries [1,2,6-11].
The objective of this study was to discriminate the genetic similarity of “Ottoman” strawberry cultivar with other local early period strawberry cultivars of Istanbul and Karadeniz Ereğli regions.
Materials and Methods
Plant material
Experiments were performed with 7 strawberry cultivars: “Aliso”, “Karaçilek”,“Tüylü”, “Ottoman” (obtained from Association of the Dissemination and Conservation of the Producers, Karadeniz Ereğli, Zonguldak, Turkey) and “Arnavutköy”, “Mangro” and “Dağ” strawberry (obtained from the garden of a family who are planting “Arnavutköy” strawberry for 100 years, Arnavutköy, Istinye, Istanbul). The samples were kept in -80 oC until analysis. There is no documentary mentioning if these cultivars were wild type or hybrids.
“Ottoman cultivar” has thick and fleshy leaves that have dark green abaxial surface and gray trichomes on the adaxial surface. This cultivar has morphological male sterile plants. The ripe fruits are in globe and bevelled globe shape. The fruits are small or middle sized. This cultivar has strong fragrance and aroma. It is classified as late season cultivar. The Ottoman strawberry cultivar is presented in (Figure 1).

Figure 1: Ottoman strawberry cultivar planted in Karadeniz Ereğli, Zonguldak
region of Turkey. (https://acatfromlondon.wordpress.com/tag/ottomanstrawberry).
“Arnavutköy” cultivar is known as the conserved cultivar of strawberry which was brought from Europe to Istanbul, Turkey. Now, there are a few families who are planting this cultivar in their small gardens in Arnavutköy, Istanbul. It grows weak and middle strength. It has a poor ability to produce stolons. The leaves are small and thin. The adaxial surfaces of the leaves are covered with silver colored trichomes. The pedicles are short and the flowers have average of 35 stamens but they are not totally functional. The ripe fruits are light red and in globe shaped. Therefore, this cultivar has significant importance for the breeding and conservation biology of the old plant species. Its small size and amazing flavor are the most characteristic features that worth conservation efforts. Fruits of “Kara” cultivar is in middle size, cone-shape and pink color. Kara, Tüylü and Mangro cultivars have male sterile specificity as Ottoman cultivar [12].
DNA isolation and RAPD-PCR analysis
DNA was extracted from the 100 mg young leaves of each strawberry cultivars by using Qiagen DNeasy Plant mini kit due to the manufacturer’s protocol. 20 oligonucleotides from Operon Technologies were used for RAPD amplifications. 10 primers of total 20 were chosen for further amplification of the DNA. The sequences of RAPD primers were given in (Table 1). RAPD-PCR reactions were carried out in a 100 μl total volume containing of PCR buffer (1X), MgCl2 (2.5 mM), dNTP (0.25 mM), primer (0.4 μM), Taq DNA polymerase (l unit) and DNA (30 ng). Amplification conditions were as follows: pre-denaturation at 95 °C for 5 min, followed by 45 cycles at 95 °C for 1 min, 35 °C for 1 min and 72 °C for 1 min. 12.5 μl of amplification products was screened in a 2% agarose gel on the Gel Doc XR+ System (BioRad, USA) [13].
Primer Code
Primer sequence (5’-3’)
OPA-02
TGCCGAGCTG
OPA-06
GGTCCCTGAC
OPB-12
CCTTGACGCA
OPB-19
ACCCCCGAAG
OPH-01
GGTCGGAGAA
OPH-06
ACGCATCGCA
OPJ-05
CTCCATGGGG
OPJ-14
CACCCGGATG
OPK-14
CCCGCTACAC
OPK-18
CCTAGTCGAG
Table 1: Primer sequences used in the RAPD-PCR.
Data analysis
The amplification bands obtained from each PCR reaction per random primers were scored as “0” or “1” scores for the absence or presence of a band, respectively in each individual sample. Similarity coefficients were calculated according to Jaccard similarity index. Sequential Agglomerative Hierarchical Non Overlapping (SAHN) clustering was applied using the Unweighted Pair Group Method with Arithmetic Averages (UPGMA). Dendrograms were plotted using NTSYS-PC Ver. 2.1 software [14].
Results
10 RAPD primers out of 20 didn’t produce any reproducible bands and the selected random primers produced 111 bands. 96 of 111 were polymorphic (86.48%). Polymorphic band numbers and polymorphism rates per perimers were given in (Table 2). The highest polymorphism rate (100%) was obtained with primers OPA06, OPB12, OPB19, OPH01, OPH06. The highest polymorphic band pattern was observed with primer OPH01. The representative amplification band patterns were given in (Figure 2). The genetic similarities between the strawberry cultivars were evaluated due to the Jaccard coefficient which is varied from 0.0327 to 0.7307 (Table 3). The average polymorhic band numbers per primer was 9.7. The RAPD bands were scored as “0” and “1” among the cultivars and UPGMA analysis was performed by using these scores. Phylogenetic analysis dendrogram was given in Figure 3. Maximum similarity was observed between Ottoman and Tüylü genotypes (73%).
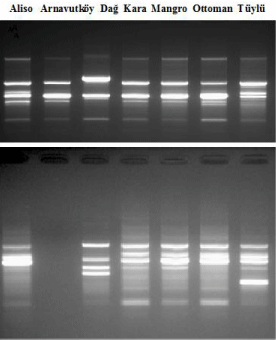
Figure 2: Representetive polymorphic band patterns obtained by RAPD
primers by OPA06 (upper) and OPH01 (lower) primers.
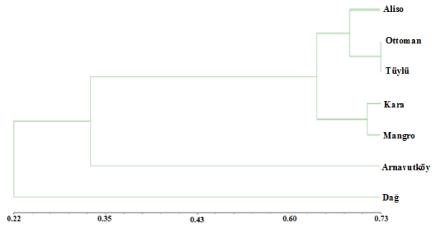
Figure 3: UPGMA Dendrogram showing the phylogenetic relationships
between strawberry cultivars.
Primer code
Polymorphic band numbers
Polymorphism rate (%)
OPA02
5
62.5
OPA06
8
100
OPB12
11
100
OPB19
12
100
OPH01
13
100
OPH06
10
100
OPJ05
7
63.6
OPJ14
11
84.2
OPK14
10
76.9
OPK18
10
83.3
Table 2: Polymorphic band numbers and polymorphism rates per random primers.
Aliso
Arnavutköy
Dağ
Kara
Mangro
Ottoman
Tüylü
Aliso
1.0000000
Arnavutköy
0.2816901
1.0000000
Dağ
0.2439024
0.0327869
1.0000000
Kara
0.6933333
0.3538462
0.2222222
1.0000000
Mangro
0.6111111
0.3750000
0.2941176
0.7121212
1.0000000
Ottoman
0.6538462
0.3432836
0.2625000
0.6800000
0.5844156
1.0000000
Tüylü
0.7215190
0.2933333
0.2857143
0.6419753
0.5844156
0.7307692
1.0000000
Table 3: Matrix of genetic similarity among strawberry cultivars based on RAPD markers calculated by Jaccard’s coefficient.
Dağ cultivar represented one cluster while the other cultivars represented the second cluster. The big upper cluster subdivided into two subclusters. Arnavutköy was classified in lower cluster individual. The upper subcluster contains Aliso, Ottoman, Tüylü, Kara and Mangro. Kara and Mangro were grouped together while Osmanli, Tüylü and Aliso were grouped in another subcluster. Aliso presented higher similarity to Tüylü with the rate of 72% whereas 65% similarity rate was observed between Aliso and Ottoman. Dağ and Arnavutköy cultivars showed the lowest genetic similarity to Ottoman with the rate of 26% and 34%. Kara cultivar showed the second highest genetic similarity rate with Ottoman (68%).
Discussion
This study aimed to discriminate the phylogenetic relationship of Ottoman and other early-period other strawberry cultivars being planted in Istanbul and Karadeniz Ereğli regions of Turkey. Strawberries were classified and identified due to their morphological and anatomical features. The results of this paper are the first evidence that identifies the genetic relation of Ottoman cultivar.
The polymorphism in genomic DNA of seven cultivars were determined by RAPD method. 10 random RAPD markers were effectively used to assess the genetic similarities between the strawberry genotypes. All the cultivars differ in size, color and flavor. The strawberry cultivars showed polymorphic bands with average rate of 87.1%. The highest level of polymorphism was observed between Dağ and Arnavutköy cultivars. Ottoman was categorized with Tüylü in a subfamily which showed the highest similarity with Aliso. Degani et al. [15] demonstrated the genetic relationship of 41 strawberry cultivars specific to USA and Canada by RAPD method. Radmann et al. (2006) discriminated the genetic similarities of the strawberry cultivars by using 26 RAPD markers with the range between 44-74%.
Milella et al. [9] determined the genetic relationship between Italian strawberry populations in Etna mountain area and their putative ancestors. They analyzed 65 selections and one cultivar “Madame Moutot” and confirmed that RAPD markers can be useful to discriminate the closely related genotypes of Fragaria x ananassa.
Kuras et al. [7] compared the applicability of RAPD and ISSR marker based methods to evaluate the genetic relationship of 24 strawberry cultivars. They evaluated both techniques efficiently to examine the genetic diversities of strawberry populations.
According to the old records Ottoman strawberry, which is indigenous to Karadeniz Ereğlisi, is believed to be improved from Arnavutköy and Kara cultivars [12]. The results supported that Kara cultivar presented high similarity rate with Ottoman cultivar (68%).
On the other hand, the genetic similarity rate between Tüylü and Ottoman cultivars (73%) is the highest level among all tested cultivars. Kara and Tüylü cultivars also showed 64% genetic similarities which both have the same male sterile characteristics with Ottoman cultivar. The morphological features and the current RAPD analysis data were both evaluated and the results indicate the necessity of further genetic analysis of Kara, Tüylü and Ottoman cultivars to highlight the phylogenetic relationship among them.
Conclusion
Our research findings clearly revealed the genetic relationship of Ottoman cultivar with other early-period strawberry varieties. Further studies must be conducted to develop in vitro micropropagation studies to protect this early-period genetic resource and to improve new industrial and agricultural usage of Ottoman strawberry cultivar.
References
- Morales RGF, Resende JTV, Faria MV, Andrade MC, Resende LV, Delattore CA, et al. Genetic similarity among strawberry cultivars assessed by RAPD and ISSR markers. Sci Agric. 2011; 68: 665-671.
- Zebrowska JI, Tyrka M. The use of RAPD markers for strawberry identification and genetic diversity studies. J Food Agric Environ. 2003; 1: 115-117.
- Atak C, Celik O, Acik L. Genetic analysis of Rhododendron mutants using Random Amplified Polymorphic DNA (RAPD). Pak J Bot. 2011; 43: 1173-1182.
- Catan R, Mitoi M, Ion R. The RAPD techniques used to assess the genetic diversity in Draba dornei, a critically endangered plant species. Adv Biosci Biotech. 2013; 4: 164-169.
- Karimi HR, Bagheriyan S, Esmaelizadeh M, Estaji A. Genetic relationships among melon using RAPD markers. Int J Veg Sci. 2016; 22: 200-208.
- Graham J, Mcnicol RJ, Mcnicol JW. A comparision of methods for the estimation of genetic diversity in strawberry cultivars. Theor Appl Genet. 1996; 93: 402-406.
- Kuras A, Korbin M, Zurawicz E. Comparison of suitability of RAPD and ISSR techniques for determination of strawberry (Fragaria × ananassa Duch) relationship. Plant Cell Tiss Org Culture. 2004; 79: 189-193.
- Sugimoto T, Tamaki K, Matsumoto J, Mamoto Y, Shiwaku K, Watanabe K. Detection of RAPD markers linked linked to the everbearing gene in japanese cultivated strawberry. Plant Breed. 2005; 124: 498-501.
- Milella L, Saluzzi D, Lapelosa M, Bertino G, Spada P, Greco I, et al. Relationships between an Italian strawberry ecotype and its ancestor using RAPD markers. Genetic Res Crop Evol. 2006; 53: 1715-1720.
- Radmann EB, Bianchi VJ, Oliveira RP, Fachinello JC. Characterization and genetic diversity of strawberry cultivars. Hortic Brasil. 2006; 26: 84-87.
- Kaur KS, Sharma R, Kumar R, Sharma SK. Molecular characterization and genetic diversity in fragaria genotypes asrevealed by Randomly Amplified DNA Polymorphisms (RAPDS). ISHS Acta Horculturae 696: VII International Symposium on Temperate Zone Fruits in the Tropics and Subtropics- Part Two. 2005; 135-142.
- Yilmaz H. Çilek, Hasad Yayincilik. 2009.
- Maniatis T, Fitsch EF, Sambrook J. In molecular cloning a laboratory manual. Cold Spring, Harbor, New York. 1982.
- Rohlf FJ. Comparative methods for the analysis of continuous variables: geometric interpretations. Evol. 2001; 55: 2143-2160.
- Degani C, Rowland LJ, Levi A, Hortynski JA, Galletta GJ. DNA fingerprinting of strawberry (Fragariaxananassa) cultivars using Randomly Amplified Polymorphic DNA (RAPD) markers. Euphytica. 2008; 102: 247-253.
