
Research Article
Austin Biomark Diagn. 2014;1(2): 5.
Genetic Variations of Mitochondrial Cytochrome B and Breast Cancer
Gazi Nurun Nahar Sultana1*, ADA Shahinuzzaman2, Rokeya Begum1, Iffat Jahan1 and M Mizanur Rahman3
1Centre for Advanced Research in Sciences, University of Dhaka, Bangladesh
2Bangladesh Council of Scientific and Industrial Research (BCSIR), Bangladesh
3National Institute of Cancer Research and Hospital, Bangladesh
*Corresponding author: Gazi Nurun Nahar Sultana, Centre for Advanced Research in Sciences (CARS), University of Dhaka, Dhaka-1000, Bangladesh.
Received: November 05, 2014; Accepted: December 09, 2014; Published: December 11, 2014
Abstract
The role of mitochondria encoded cytochrome b (cyt b) gene mutations has been drawn a great interest to understand the function of this gene in various cancer including breast cancer. Although most cancer cells harbor somatic mutations in mitochondrial DNA (mtDNA) including cytochrome b gene, the question of whether such mutations in cyt b contribute to the promotion of breast cancer remains obscured. In this study, we investigated the frequency of mutations in cyt b gene in forty (n = 40) breast cancer patients of Bangladesh. The results were compared with forty eight age matched control samples (n = 48) from our database. Nine (9) different sequence variations were found on cyt b genein cancer samples. Detail study of cyt b gene of forty eight (n = 48) controls found eighteen (n = 18) different variations, which demonstrates the high variability of cyt b sequence. Three of these eighteen mutations 15055 (G>A), 15301 (G>A), and 16326 (A>G) are also found in cancer patients. Two mutations in breast cancer patients were homoplasmic and identified as major percentage compared to control samples. Two mutations have been identified at np 14766 (C>T) 100%, followed by 14783 (T>C) in 40%breast cancer samples. Two other mutations at np15479 (T>C) and 15497 (G>A) have been reported in 2% breast cancer patients but absent from control samples. Three phylogenetic variations at np15055 (T>C), 15301 (G>A) and 15326 (A>G) have been found in 18% cancer patients respectively but also found in control samples. We have identified two insertion polymorphisms at 15970 (A-ins) and 15980 (T-ins) in 2% cancer samples. Thus, we hypothesized that mutation at np 14766 (C>T) 100% in cyt b gene may cause defective assembly and function of complex- III which thus hampers ATP production in cancer cells in addition with others mechanisms of breast cancer development. As a result, cancer cells are forced to use glycolytic pathways for ATP production. Alternatively two mutations14766 (C>T) and 15326 (G>A) lead to frame shift in amino acids (T7I) and (T194A) of translated protein. However report from the software analysis suggests that this (T7I) change might not be tolerated in protein function with some deleterious effect.
Keywords: Breast cancer; Mitochondrial cytochrome; mtDNA; OXPHOS
Introduction
The risk of mitochondrial DNA (mtDNA) mutations with breast cancer is just in the beginning of understanding. Not much investigation has been done related to mitochondrial cytochrome b gene mutation in breast cancer patients. Although there are several reports on cyt b gene mutations in different cancer but functional effect of these mutations in tumor development is yet to be reveled [1,2]. Mitochondria play a vital role for regulation of Oxidative Phosphorylation System (OXPHOS) producing cellular currency called ATP. The OXPOS system is composed of five complexes (I-V) and some of them are encoded by mitochondrial DNA (mtDNA) and others by nuclear DNA [3]. Only cyt b of mitochondria plays a vital role for the assembly and function of complex-III, and together with Cytochrome c1 and iron-sulfur protein, it forms the catalytic core of the enzymes [4]. In particular, mutations in the mtDNA encoded cyt bgene are associated with combine complex I+III deficiency or with only complex III deficiency [5]. As a result functional defect in the OXPHOS system may force to impede electron flow down the electron transport chain could increase ROS production and contribute to cancer [6,7]. It was shown in other study that over expression of cyt b generated increase ROS accompanied by increase oxygen consumption and lactate production. Cyt b over expression induced significant tumor growth in vitro and in vivo by triggering significant cell cycle progression through up regulation of nuclear factor NF?B2 signaling pathway and may set up cell for further cell cycle progression, invasion and inhibition of apoptosis [8,9]. In this study, we examined the hypothesis that accumulation of mutation in cyt b gene might be an additional risk factor for the promotion of tumor growth in breast cancer.
Materials and Methods
Sample collection
All approval has been taken from the local ethical committee of Bangladesh Medical Research Council (BMRC) and University of Dhaka. Forty (n = 40) breast cancer patients from Dhaka Medical College Hospital, Dhaka and National Institute of Cancer Research Hospital (NICRH), Dhaka, Bangladesh were included for the study in 2010-2012. We have collected personal history of those patients who were diagnosed as breast cancer. Blood and tumor tissue samples were collected from 37 patients during surgical operation before chemotherapy. Rest of the 3 samples was collected from patients who already had chemotherapy before surgery. A healthy cohort of 48 age matched female individuals from mainstream population was also included. All two populations share the same ethnicity and nationality and reside in Bangladesh. Subjects with breast cancer were interviewed. Blood samples 3-5 mL was collected in EDTA coated tubes from breast cancer patients who visited the hospital for treatment and tissue samples were collected in screw capped polypropylene vials during surgery. The samples were transported to laboratory and blood samples kept at -20°C and tissue samples at -40°C until analyzed.
DNA isolation, PCR and sequencing
Cancer and normal blood DNA was isolated by standard proteinase K treatment followed by phenol/ chloroform/ isoamyl alcohol extraction. Tissue samples were isolated by DNA extraction kit purchased from (Qiagen, Turn berry LaneValencia, California). DNA was precipitated with 0.3 M sodium acetate (pH 5.2) in 70% ethanol at -20°C overnight and suspended in Tris-EDTA (TE) buffer (pH 8.0). DNA quantification was performed by taking absorbance at 260 nm and visualized by 0.8% agarose gel electrophoresis [10]. Partial mitochondrial genome was amplified using two sets of primers Table 1 and the resultant amplicon were checked in 2% agarose gel electrophoresis [11]. Distinct PCR bands were observed for different primers amplifying Cytochrome b gene, along with diluted 1 Kb Plus DNA ladder purchased from life Technologies, USA (Cat no. 10787-018). 20 μl PCR reaction contained 10-20 ng DNA and 0.5 μM primers, 0.2 mM each of deoxynucleotide triphosphate (dNTP), 1U of TaqMan™ DNA Polymerase (Applied Biosystems, USA) and 2.5 mM MgCl2. The PCR amplification of specific regions of mtDNA was performed on the basis of following cycling conditions: initial denaturing at 95°C for 5 min followed by 94°C for 30 sec, 58°C for 30 sec, and 72°C for 2 min for 35 cycles and final extension step at 72°C for 7 min. In Sequencing PCR, the ABI-prism Big Dye Terminator V 3.1 containing ampliTaq polymerase, dye terminators (fluorescent label), deoxynucleotide triphosphate, magnesium chloride, was used for direct sequencing of PCR product for specific primers (forward/ reverse primer). The sequencing PCR was performed on the basis of following cycling conditions: initial denaturing at 95°C for 1 min followed by 94°C for 10 sec, 55°C for 30 sec, and 60°C for 4 min for 35 cycles.
Primer ID
Primer Sequence
3' position
Length
Overlap
1F
5' GCATAATTAAACTTTACTTC 3'
14000
-
-
1R
5' AGAATATTGAGGCGCCATTG 3'
14998
938
206
2F
5' TGAAACTTCGGCTCACTCCT 3'
14856
-
-
2R
5' AGCTTTGGGTGATAATGGTG 3'
15978
1162
180
Table 1: Primer used for Cyt B gene amplification.
MtDNA sequence analysis
The purified sequencing PCR products were analyzed by electrophoresis in the ABI-Prism 3130 Genetic Analyzer (Applied Biosystems, USA). The sequence patterns were observed and edited by using Mac-based software (Auto Assembler V 3.0) and BioEdit Sequence Alignment Editor V 7.0.9.0 (https://www.mbio.ncsu.edu/ bioedit/bioedit.html). The sequences were aligned by using bl2seq tool of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared with the revised Cambridge Reference Sequence, rCRS (NCBI Reference Sequence: NC_012920.1) [12]. MtDNA polymorphisms were compared with the mitochondrial genome database of world population by using Mitomap (www.mitomap.org).
Statistical analysis
Two tailed non-directional Fisher-Irwin (Fisher's exact test) has been used to verify the mutation probability difference in cancer and healthy (control) populations [13] To confirm the result of this common test in the light of low expected number in the healthy population Yates's chi and un-corrected chi squared test ('N-1' chi squared test) have been used as expected to give relatively low Type I error. The analysis was performed as previously described [14]. To further understand the significance of two polymorphism related risk factor for unfavorable outcomes (odds ratio, relative risk, difference in proportions, absolute and relative reduction in risk, number needed to treat) and of the effectiveness of a diagnostic criteria (sensitivity, specificity, positive and negative predictive values, positive and negative likelihood ratios, diagnostic and error odds ratios) has been performed [15]. The p value <0.05 is significant. The parameters, as well as the confidence intervals for the estimated parameters are computed by standard methods [16].
Results
We have identified nine different mutations in cytochrome b gene (Table 2) of which the most frequent mutations were found at np14766 (C>T) and np 14783 (T>C), which occurred in 100% and 40% of breast cancer patients (Figure 1). The mutation 14766 (C>T) has high sensitivity (1.00, 95% confidence interval 0.932-1.000, and odd ration 230.00) and this mutation 14766 (C>T) is statistically significant (P = 0.0001). For second mutation at np 14783 (sensitivity 0.450, 95% confidence interval 21.8678 to 17.5987, and odd ration: 6.7333) which is also statistically significant (P = 0.0023).Two other additional mutations at np 15479 (T>C) and 15497 (G>A) have been reported in 2% cancer patients.Three phylogenetic variations at nt 15055 (T>C), 15301 (G>A) and 15326 (A>G) have been found in (18%) cancer patients as well as in 10%, 39% and 80% control samples respectively. The base substitution at 14766 (C>T) in mitochondrial cytochrome b gene leads to a shift in amino acid (T7I) of translated protein which converts the amino acid threonine to isoleucine, 14783 (T>C) converts leucine to leucine (L13L) and 15326 (T>C) converts amino acid tyrosine tohistidine(T194A). We identified two insertion polymorphisms at 15970 (A-ins) and 15980 (T-ins) in five samples whereas no such changes were observed in control. Here we also reported the eighteen different variations in 48 control healthy individuals one of which is newly reported (Table 3).
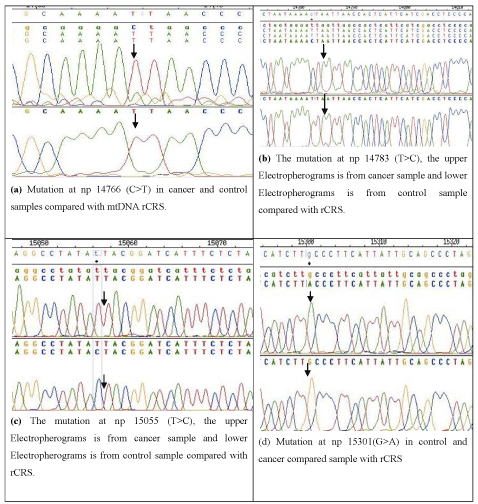
Figure 1: Shows Electropherograms of four mutations both from control and cancer samples compared with rCRS.
Position
Mutation
Codon No
AA changes
14766
C>T
7
Thr-Iso
14783
T>C
13
Leu-Leu
15055
T>C
103
Tyr-Tyr
15301
G>A
185
Leu-Leu
15326
A>G
194
Thr-Ala
15479
T>C
245
Phe-Leu
15497
G>A
251
Gly-Ser
15970
A-ins
new
-
15980
T-ins
new
-
Table 2: Nine mutations identifies in mtDNA cytochrome b gene in breast cancer of Bangladeshi.
Position
Mutation
Codon
AA changes
Codon position
14783
T>C
13
L-L
1
15040
C>T
98
L-L
3
15043
G>A
102
??
4
15071
T>A
109
Y-H
1
15301
G>A
185
L-L
3
15583
A>G
-
-
-
15326
A>G
94
Thr-Ala
1
15055
T>C
103
Tyr-Tyr
3
15974
A>G
-
-
tRNA
15691
A>G
315
M-M
3
15607
A>G
287
Lys-Lys
3
15379
C>T
311
Lys-Lys
3
15896
A>G
-
-
tRNA
15564
A>G (new)
-
-
-
15773
G>A
343
Val-Met
1
15530
T>C
262
Leu-Leu
1
15440
T>C
-
Lys-Lys
-
15221
G>A
159
Asp-Asn
1
15601
T>C
285
Pro-Pro
3
Table 3: Eighteen different variations identifies in mtDNA Cytochrome b gene in healthy control women of Bangladesh.
Discussion
Carcinogenesis is referred as a multistep process [17], through which a normal cell both genetically and metabolically evolves spontaneously over years or decades to develop into clinical cancer. Being the power house of the cell, the mitochondrion is one of the key centers where such diverse alteration take place extending from the range of reactive oxygen species generation to the decrease protein level of Cytochrome b subunit of F1 of ATPase [18] leading to altered ATPase function. We believe that defining mtDNA polymorphisms and/or mutations patterns in selected type of cancers, including breast cancer may help to understand the basic biochemical mechanisms involved in the induction of cell transformation and indicate the potential role of mtDNA in cancer progression.
The great variability of the human mitochondrial DNA (mtDNA) sequence induces many difficulties in the search for its deleterious mutations [19]. We illustrate these pitfalls by the analysis of the cytochrome b gene of 40 breast cancer patients. Nine different sequence variations were found, of which two are new mutation. Extensive analysis of the cytochrome b gene of 48 controls samples provided 18 supplementary mutations where one mutation is new, thus further demonstrating the high variability of the cytochrome b sequence. We evaluated the functional relevance of five mutations out of 9 using indirect criteria such as the nature of the mutation, and its frequency in case-controls. Homoplasmic mutations indicating that the mutant genome was dominant at the intracellular and intercellular levels [20]. The cytochrome b of mitochondrion is a component of the ubiquinol-cytochrome c reductase complex (Complex III or cytochrome b-c1 complex), which is a respiratory chain that generates an electrochemical potential coupled at ATP synthesis [21]. The mutation at np14766 (C>T) was observed in 100% samples and is highly significant (P = 0.0001) the second mutation 14783 (T>C) was observed in 40% patients and is also statistically significant (P = 0.0023). The third mutation (15326 (A>G) have been identified in 18% cases which is also present in 80% control samples. The bioinformatics analysis confirmed the intolerance of T7I but tolerance of L13L and T194A alteration in Cytochrome b protein.Our result is supported by other study which elucidates the main effect of mtDNA variants on the risk of developing breast cancer through understanding of gene to gene interaction [22].
Such mutations might influence function of mRNA and/or mtDNA regulatory regions by yet unidentified mechanisms and provide a functional advantage for the cancer cell [23]. In addition, it is more possible that the mutation 15326 (A>G) may not have any contribution to breast cancer risk association but is selected in two different populations through evolution and migration, that's why this might have become prevalent mutations in both population in process of natural selection [24]. The other mutation 14766 (C>T) lead to change in amino acid (T7I) in cytochrome b protein. Using the same analysis with 'Sorting Intolerance From Tolerance' (SIFT) and PMut, it was estimated that this amino acid change (T7I) will not be tolerated and L13L, T194A will be tolerated. Thus it is concluded that mutation at np 14766 (C>T) might be a risk factor for developing breast cancer in combination with other genetic and epigenetic risk factor but 14783 (T>C) and 15326 (A>G) is an altered allele with no possible role in malignancy. Moreover, the role of mtDNA protein in the development of breast cancer has been studied in recent years. The potential biomarker of the mitochondrial complex I subunit has been found significantly associated with breast cancer aggressiveness [25-28].
The single mutation cannot be attributed as a major risk factor for breast cancer though we studied both tumor samples and body fluids for cyt b gene. Therefore suchsingle mutation can be considered as marker for breast cancer identification with increased sample size of primary breast cancer in combination with our previous study [29]. Future research to address the functional aspects of mtDNA mutations in cancer development and therapeutic response is likely to be fruitful and have significant clinical implications in the prevention and treatment of cancer.
Acknowledgement
The authors thank to all patients for cooperating during sample collection. The authors are also thankful to control blood donors. Authors are grateful to Sharif Moshiuzzaman for DNA extraction and purification.
References
- Dasgupta S, Hoque MO, Upadhyay S, Sidransky D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008; 68: 700-706.
- Croteau DL, Bohr VA. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J Biol Chem. 1997; 272: 25409-25412.
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008; 32: 529-539.
- Burgart LJ, Zheng J, Shu Q, Strickler JG, Shibata D. Somatic mitochondrial mutation in gastric cancer. Am J Pathol. 1995; 147: 1105-1111.
- Yeh JJ, Lunetta KL, van Orsouw NJ, Moore FD Jr, Mutter GL, Vijg J, et al. Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene. 2000; 19: 2060-2066.
- Richter C, Gogvadze V, Laffranchi R, Schlapbach R, Schweizer M, Suter M, et al. Oxidants in mitochondria: from physiology to diseases. Biochim Biophys Acta. 1995; 1271: 67-74.
- Richard SM, Bailliet G, Paez GL, Bianchi MS, Peltomaki P, Bianchi NO. Nuclear and mitochondrial genome instability in human breast cancer. Cancer Res. 2000; 60: 4231-4237.
- Ravi R, Bedi GC, Engstrom LW, Zeng Q, Mookerjee B, Gelinas C, et al. Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-kappaB. Nat Cell Biol. 2001; 3: 409-416.
- Franco AV, Zhang XD, Van Berkel E, Sanders JE, Zhang XY, Thomas WD, et al. The role of NF-kappa B in TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of melanoma cells. J Immunol. 2001; 166: 5337-5345.
- Sambrook J, Russell D. Molecular Cloning: A laboratory Manual 3rd edition. 2000.
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998; 26: 967-973.
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981; 290: 457-465.
- Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. Boca Raton: Chapman & Hall. 2007; 29: 12.
- Campbell I. Chi-squared and Fisher-Irwin tests of two-by-two tables with small sample recommendations. Stat Med. 2007; 26: 3661-3675.
- Sackett DL, Deeks JJ, Altman DG. Down with odds ratios! Evid Based Med. 1996; 1: 164-166.
- Newcomb RG. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Statistics in Medicine. 1998; 17: 857-872.
- Pelicano H, Martin DS, Xu RH, Huang P. Glycolysis inhibition for anticancer treatment. Oncogene. 2006; 25: 4633-4646.
- Voo KS, Zeng G, Mu JB, Zhou J, Su XZ, Wang RF. CD4+ T-cell response to mitochondrial cytochrome B in human melanoma. Cancer Res. 2006; 66: 5919-5926.
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005; 39: 359-407.
- Duesberg P, Li R. Multistep carcinogenesis: a chain reaction of aneuploidizations. Cell Cycle. 2003; 2: 202-210.
- Emil Y, Anu S. Trends in Molecular Medicine: Mechanisms of mitochondrial diseases. Annals of Med. 2011; 23: 1-9.
- Ma Y, Bai RK, Trieu R, Wong LJ. Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta. 2010; 1797: 29-37.
- Covarrubias D, Bai RK, Wong LJ, Leal SM. Mitochondrial DNA variant interactions modify breast cancer risk. J Hum Genet. 2008; 53: 924-928.
- Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, et al. Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res. 2001; 61: 7623-7626.
- Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002; 62: 972-976.
- Suhane S, Berel D, Ramanujan VK. Biomarker signatures of mitochondrial NDUFS3 in invasive breast carcinoma. Biochem Biophys Res Commun. 2011; 412: 590-595.
- Sultana GN, Rahman A, Shahinuzzaman AD, Begum RA, Hossain CF. Mitochondrial DNA mutations---candidate biomarkers for breast cancer diagnosis in Bangladesh. Chin J Cancer. 2012; 31: 449-454.
- Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007; 67: 4687-4694.
- Sultana GNN, Rahman A, Karim MM, A. D. A. Shahinuzzaman, Rokeya Begum, Rowsan Ara Begum. Breast cancer risk associated mitochondrial NADH dehydrogenase subunit3(ND3) polymorphisms (G10398A and T10400C) in Bangladeshiwomen. J Med Genet. 2011; 3: 131-135.