Review Article
Austin J Biomed Eng. 2014;1(4): 1017.
Profiling Protein-Protein Interactions and Protein Structures Using Chemical Cross-linking and Mass Spectrometry
Shengjie Bian and Saiful M Chowdhury*
Department of Chemistry and Biochemistry, University of Texas at Arlington, USA
*Corresponding author: :Saiful M Chowdhury, Department of Chemistry and Biochemistry, University of Texas at Arlington, 700 Planetarium Place, Rm 130, Arlington, TX, USA.
Received: July 02, 2014; Accepted: Aug 04, 2014; Published: Aug 06, 2014
Abstract
Protein cross-linking strategies coupled to mass spectrometry is a powerful tool for studying protein structures, protein surface topologies and protein-protein interactions. There are more than one hundred chemical crosslinkers, and some of them are already commercially available. However, the current researches require new and more effective crosslinkers and more intelligent data analysis software tools. Here, we describe some new features of crosslinkers such as cleavable cross-linkers that facilitate detection by mass spectrometry, affinity crosslinkers using click-chemistry that facilitate separation and enrichment of cross-linked peptides, and crosslinkers with the combined features. In addition, the evolution in software remarkably improves the data analysis efficiency and makes it possible to perform large-scale identification of cross-linked peptides. The advances in protein cross-linking analysis software are also addressed.
Keywords: Mass spectrometry; Chemical crosslinker; Protein cross-linking; Proteomics
Abbreviations
MS/MS: Tandem MS; LC: Liquid Chromatography; CID: Collision Induced Dissociation; ETD: Electron Transfer Dissociation; NHS: N-hydroxysuccinimide; DSS: Disuccinimidyl Suberate; BS3: Bis[sulfosuccinimidyl]Suberate; CLIP: Click-enabled Linker for Interacting Proteins; PIR: Protein Interaction Reporter.
Introduction
Chemical cross-linking combined with mass spectrometry can be a powerful approach for the identification of protein-protein interactions and providing information on protein structures [1-3]. This technique is emerging as an alternative to traditional structural biological methods such as time-consuming crystallography-based X-ray analyses and NMR-based approaches. The current chemical crosslinkers provide the feasibility to study proteins structures and interactions. However, the heterogeneity and low abundance of the cross-linked products continue to pose enormous challenges for large-scale biological application of cross-linking approaches [4]. Moreover, the huge amount of data collected as well as the theoretical combinatorial complexity inherent to the cross-linking process, has encouraged the scientists to develop specialized software tools to analyze mass spectra, to mine databases, and to generate pertinent structural information. In this review, we address several important issues that are related to the studies of protein structures and proteinprotein interactions by cross-linking coupled to mass spectrometry analysis. Furthermore, the development of novel cross-linking reagents with some new features in recent years to optimize crosslinking strategies for mass spectrometry analysis is also described. Currently our laboratory focuses on the development of novel CID-/ ETD-cleavable crosslinkers to facilitate mass spectrometry detection of cross-linked peptides as well as their application to decipher innate immune signaling pathways of Toll-like Receptors (TLRs).
Protein cross-linking and new features of chemical crosslinkers
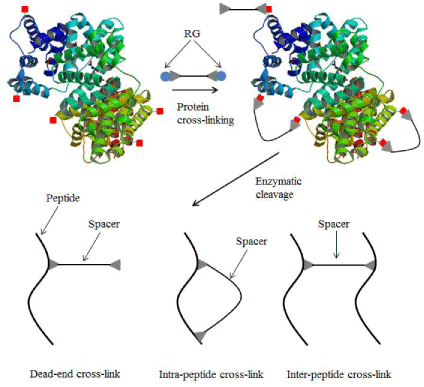
Cross-linking is the process of formation of covalent bonds that links one molecule to another. In biological studies, it refers to the use of a probe to link two interacting proteins in order to study protein structures and protein-protein interactions. It is used to determine the domains of proteins which are close to each other to form covalent linkage in a protein complex, and to determine the sites where the reaction occurs. In relation to the size of the crosslinkers, the data collected can be viewed as a set of distance constraints providing clues on protein structures and the topologies of multi-protein complex [5]. The major challenges in protein interaction studies with chemical cross-linking arises from the complexity of intra-, inter-, and dead-end cross-linked peptide mixtures (Figure 1)[6]. Although, there is remarkable advancement in mass spectrometry instruments in recent years but there is still a great need for effective crosslinker with innovative features as well as user-friendly data analysis software to advance this technology for large-scale applications, such as deciphering the systems-level protein signaling networks.
More than one hundred of cross-linkers have been described in the literature. Most often the targets of cross-linking functional groups are the strong nucleophiles such as the sulfhydryl and amine groups of cysteine and lysine side chains. There are varieties of crosslinkers that employ N-hydroxysuccinimide (NHS) esters for reacting with amine groups and N-maleimide for reacting with thiols. For example, disuccinimidyl suberate (DSS) and its more water-soluble sulfonate analogue bis[sulfosuccinimidyl] suberate (BS3) is a commonly used crosslinker which reacts easily with the amine groups of the proteins. In recent years, some new crosslinkers are emerging, which have critical features, such as isotopic coding, cleavability, affinity groups, new reactive groups, and compatibility with mass spectrometry [7]. These chemical crosslinkers with new features can assist to facilitate the data acquisition and analysis by mass spectrometry.
Figure 1: Schematic diagram of protein cross-linking and three types of cross-links including dead-end. Cross-link, intra-peptide cross-link and interpeptide cross-link (RG - Reactive groups).
Cleavable crosslinkers and click-based enrichment
The introduction of a cleavable bond within the crosslinker structure simplifies the identification of cross-linked peptides and data analysis after tandem mass spectrometry analysis [8]. The specific cleavable bond can be cleaved either chemically or by fragmentation with tandem mass spectrometry systems (for e.g. under CID or ETD condition), and results in unique signatures in mass spectra, which can greatly simplify data analysis. A widely used mass spectrometrycleavable cross-linking strategy is Protein Interaction Reporter (PIR) technology, which was successfully applied for in vivo identification of protein–protein interactions as well as actual regions of the interacting proteins that share close proximity while present within cells [4,6]. Moreover, the purification of cross-linked peptides can be facilitated when the crosslinker contains specific affinity group. The typical affinity-based approach used in crosslinkers is the biotinavidin affinity capture strategies. However, the drawback of affinity based crosslinkers is that they need to be custom synthesized and are generally more bulky than conventional reagents. This may affect their reactivity because of steric hindrance, and the accuracy of spatial constraints is reduced [9]. Thus, multifunctional but compact crosslinkers must be addressed.
A compact crosslinker was reported by Chowdhury et al. with two distinct features [1]. The crosslinker consists of an alkyne tag which enabled enrichment of the cross-linked peptides after proteolytic cleavage using alkyne-azido click chemistry. A small molecule detection tag (NO2) in the crosslinker enabled the detection of neutral loss of this small NO2 moiety as a secondary means of detecting cross-linked peptides in MS/MS analyses, which provided additional confidence in peptide identifications. The CLIP (clickenabled linker for interacting proteins) technique facilitates both peptide enrichment and mass spectrometry data acquisition. Using CLIP, cross-linked ubiquitin peptides were successfully enriched from complex samples and the automated identification of crosslinked peptides was improved with both CID-MS/MS (collisioninduced dissociation tandem mass spectrometry) and ETD-MS/MS (electron transfer dissociation) modes.
New computer software to decipher acquired MS data from cross-linked peptides
The analysis of MS data from cross-linking experiments with traditional non- cleavable crosslinkers is still a challenge because of the large number of possible combinations that have to be considered in the search algorithm. Some publicly available processing tools, such as xComb and xQuest have been developed for use with standard proteome search engine to simplify the identification of crosslinked peptides [10,11]. Other individualized bioinformatics tools were also developed to do more specialized analysis of cross-linked data [12-14]. Nevertheless, the identification of cross-links from biological protein complex is still challenging and more powerful and individualized bioinformatics tools need to be developed for the complicated calculation and assignment of the big data set from the cross-linked mass spectrometry data. Currently, there is no effectivesoftware available to study CID-cleavable cross-linking data from large-scale application.
Du et al. [15] established a comprehensive data analysis platform, Xlink-Identifier, which has been developed to support label-free analyses of data from traditional non-cleavable crosslinkers. It can identify inter-peptide, intra-peptide, and dead-end cross-links as well as underivatized peptides. The software streamlines data preprocessing, peptide scoring, and visualization and provides an overall data analysis strategy for studying protein-protein interactions and protein structures using mass spectrometry. Xlink-Identifier offers the potential to perform large-scale identifications of proteinprotein interactions using tandem mass spectrometry.
Conclusions and Perspectives
Over the past two decades, mass spectrometry (MS) analysis of peptides and proteins has evolved dramatically. With the aid of chemical crosslinkers coupled to mass spectrometry, the structure information of proteins and protein-protein interactions can be studied in an accurate and efficient way. However, some technical limits still exist, for e.g. the current database is not integrated with cross-link information and special software has to be used for crosslinking analysis. The future demands to advance this research field require more intelligent analytical software tools and more versatile chemical crosslinkers with innovative features, which can identify protein signaling networks with high confidence.
References
- Chowdhury SM, Du X, Tolic N, Wu S, Moore RJ, Mayer MU, et al. Identification of cross-linked peptides after click-based enrichment using sequential collision-induced dissociation and electron transfer dissociation tandem mass spectrometry. See comment in PubMed Commons below Anal Chem. 2009; 81: 5524-5532.
- Pettelkau J, Thondorf I, Theisgen S, Lilie H, Schröder T, Arlt C, et al. Structural analysis of guanylyl cyclase-activating protein-2 (GCAP-2) homodimer by stable isotope-labeling, chemical cross-linking, and mass spectrometry. See comment in PubMed Commons below J Am Soc Mass Spectrom. 2013; 24: 1969-1979.
- Müller MQ, Dreiocker F, Ihling CH, Schäfer M, Sinz A. Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. See comment in PubMed Commons below Anal Chem. 2010; 82: 6958-6968.
- Tang X, Bruce JE. A new cross-linking strategy: protein interaction reporter (PIR) technology for protein-protein interaction studies. See comment in PubMed Commons below Mol Biosyst. 2010; 6: 939-947.
- Paramelle D, Miralles G, Subra G, Martinez J. Chemical cross-linkers for protein structure studies by mass spectrometry. See comment in PubMed Commons below Proteomics. 2013; 13: 438-456.
- Chowdhury SM, Munske GR, Tang X, Bruce JE. Collisionally activated dissociation and electron capture dissociation of several mass spectrometry-identifiable chemical cross-linkers. See comment in PubMed Commons below Anal Chem. 2006; 78: 8183-8193.
- Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. See comment in PubMed Commons below Mass Spectrom Rev. 2010; 29: 862-876.
- Soderblom EJ, Bobay BG, Cavanagh J, Goshe MB. Tandem mass spectrometry acquisition approaches to enhance identification of protein-protein interactions using low-energy collision-induced dissociative chemical crosslinking reagents. See comment in PubMed Commons below Rapid Commun Mass Spectrom. 2007; 21: 3395-3408.
- Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, et al. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. See comment in PubMed Commons below Mol Cell Proteomics. 2010; 9: 1634-1649.
- Rinner O, Seebacher J, Walzthoeni T, Mueller LN, Beck M, Schmidt A, et al. Identification of cross-linked peptides from large sequence databases. See comment in PubMed Commons below Nat Methods. 2008; 5: 315-318.
- Panchaud A, Singh P, Shaffer SA, Goodlett DR. xComb: a cross-linked peptide database approach to protein-protein interaction analysis. See comment in PubMed Commons below J Proteome Res. 2010; 9: 2508-2515.
- Chu F, Baker PR, Burlingame AL, Chalkley RJ. Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. See comment in PubMed Commons below Mol Cell Proteomics. 2010; 9: 25-31.
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, et al. Identification of cross-linked peptides from complex samples. See comment in PubMed Commons below Nat Methods. 2012; 9: 904-906.
- Nadeau OW, Wyckoff GJ, Paschall JE, Artigues A, Sage J, Villar MT, et al. CrossSearch, a user-friendly search engine for detecting chemically cross-linked peptides in conjugated proteins. See comment in PubMed Commons below Mol Cell Proteomics. 2008; 7: 739-749.
- Du X, Chowdhury SM, Manes NP, Wu S, Mayer MU, Adkins JN, et al. Xlink-identifier: an automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. See comment in PubMed Commons below J Proteome Res. 2011; 10: 923-931.