
Review Article
Austin Biom and Biostat. 2016; 3(1): 1030.
Big Data Science and its Applications in Healthcare and Medical Research: Challenges and Opportunities
Liang Y¹* and Kelemen A²
¹Department of Family and Community Health, University of Maryland, Baltimore, USA
²Department of Organizational Systems and Adult Health, University of Maryland, USA
*Corresponding author: Liang Y, Department of Family and Community Health, University of Maryland, Baltimore, MD 21201, USA
Received: May 06, 2016; Accepted: June 02, 2016; Published: June 09, 2016
Abstract
Recently, Big Data science has been a hot topic in the scientific, industrial and the business worlds. The healthcare and biomedical sciences have rapidly become data-intensive as investigators are generating and using large, complex, high dimensional and diverse domain specific datasets. This paper provides a general survey of recent progress and advances in Big Data science, healthcare, and biomedical research. Big Data science impacts, important features, infrastructures, and basic and advanced analytical tools are presented in detail. Additionally, various challenges, debates, and opportunities inside this quickly emerging scientific field are explored. The human genome and omics research, one of the most promising medical and health areas as an example and application of Big Data science, is discussed to demonstrate how the adaptive advanced computational analytical tools could be utilized for transforming millions of data points into predictions and diagnostics for precision medicine and personalized healthcare with better patient outcomes.
Keywords: Big data science; Big data infrastructure; Advanced analytics; Human genomics and OMICS; Precision medicine; Healthcare
Introduction
The big data impact and potentials in healthcare and medical sciences
Big Data is more than a decade old term that became very popular recently in life sciences and other fields. The healthcare industry has always been a large generator of biomedical data, with the U.S. healthcare system expected to reach the zettabyte (10²¹) scale from electronic health records, scientific instruments, clinical decision support systems, or even research articles in medical journals [1- 3]. Biomedical enterprises including the fields of human genomics (e.g., NIH 1000 Genome project), medical imaging (e.g., BRAIN initiative), the growth of mHealth, telehealth, and telemedicine, have generated trillions of data points resulting from the recent advances in biotechnology and advent of new computing sources (such as cloud) [4-14]. Big Data and its practices in health or medical science become even more prominent due to new social arenas/media and networks (such as Facebook and Twitter), sensory/digital technology, and mobile devices with smartphone apps and personal sensor health data with real time digital data accumulations [15,16].
The National Institutes of Health announced the Big Data to Knowledge (BD2K) Initiative with its long-term goals in 2014. As an important exemplar, NIH recently announced the “Precision Medicine Initiative”, which intends to assemble a longitudinal “cohort” of 1 million Americans, and characterize extensively with cell populations, proteins, metabolites, RNA, DNA and whole genome sequencing along with behavioral data; all linked to electronic health records, and eventually develop genetically guided therapy in the personalized and precision medicine for better preventive solution, early detections and treatment of common complex diseases [14,17- 21]. In the healthcare public health domains, AHRQ and Patient Centered Outcome Research (PCORI) have launched the PCORnet initiative to support an effective, sustainable national research infrastructure that advances data collection from very large study populations, shares and uses of electronic health data in Comparative Effectiveness Research (CER) and other evidence based practice/ medicine research [22-25].
For the educational standard, Big Data are gradually driving higher education from data poor to data rich domain, from hypothesis driven to data driven, and the movements of the online or web based educations as “Wind Tunnels” promote more students getting involved in learning Big Data science worldwide. For example, at the University of London, UK, the Big Data Society forum, related journal, and the Big Data school certificate that trains next generation Big Data science researchers have been established [26-29]. Big Data science has been gradually recognized as an emerging field and discipline and could be one of the most valuable assets not only in the life sciences such as medical and healthcare, but also other domains including educational standards, government prospective, social sciences, financial industry and business opportunities [4-6,30-34]. The lessons learned from all those related domains and fields could be potentially applied to the healthcare and medical fields, e.g., from business field for the lowered cost, improved quality outcomes (fewer medical errors and readmissions), increased efficiency, productivity, effectiveness, and performance of healthcare providers and associated systems.
Big data science features and infrastructure
Big Data science refers to the massive amounts of multiple digital data sets that are captured, collected, integrated, and analyzed. The important features of Big Data include: 1) size/ scale in terms of Volume, Velocity, Variety (known as three V’s): mass of measures increased from petabytes to exabytes, zettabytes, yottabytes; 2) evolving, varied, distributed, timeliness, dynamic, not static, change with real time; 3) complexity and heterogeneity (structured, unstructured, semi-structured data); 4) data sharing and privacy [7,35-39] Due to these unique properties, in order to maximize Big Data potentials for knowledge discovery, and make it actionable and operational for better life science solutions, Big Data science infrastructure, the intelligent fundamental analytical tools, and advanced computational approaches that could conceptualize, theorize, and model the Big Data with the grounded theory method need to be established, understood and available by both Data analysts and domain researchers [40,41]. Therefore, a top layer question for Big Data scientists is what the important framework for good Big Data governance and implementation is in order to make it actionable and operational. There are four critical hierarchical domains/levels for the infrastructure of the Big Data governance [42].
First, in the software, hardware, and physical capacity domains, Big Data requires parallel-distributed architectures with a high performance multicore and clustering or cloud computing platforms that can access hundreds or even thousands of processors. The Hadoop system is an example, and is a distributed computing environment using a Map-Reduce framework. Hadoop tools and related software including HDFS distributed file systems allow for the storage, backup and computing resources for complex workloads [43-49]. Software-defined data center or software-defined network is open flow application programming to interfaces or a virtual network overlay for controlling, understanding and dealing with Big Data, which could also create agility and automation with a centrally programmable network [50,51]. Big data Script is an example of scripting language for complex big data processing pipeline, which improve the hardware abstraction and execution from wide ranges of computer architecture from laptop, to multicore servers, to cloud computing [52].
A few other examples of popular computing software include i) the open source R statistical language and related packages such as bioconductor has been well utilized in the past decades for analyzing Big genomic data [53]; ii) open source pbdR software is a series of R packages and an environment for statistical computing and programming with Big Data in R (https://r-pbd.org) [54,55]. Note that the difference between pbdR and R codes is that R system focuses on single multi-core machines for data analysis via an interactive mode such as GUI interface; while pbdR focuses on distributed memory system, where data are distributed across several processors and analyzed in a batch mode, and communications between processors are utilized in large High-Performance Computing (HPC) systems; iii) Revolution Analytics is a free and premium software and services that brings high-performance, productive, and ease-of-use to R and enables data scientists to derive greater meaning from large sets of critical data in record time; iv) Tableau Software, Tableau Desktop and Tableau Server uses visual analytics, ease-of-use approach and flexibility connecting to live data and perform visual, rapid-fire analysis.
Second, in the databases level/domain, to manage large volume unstructured (e.g., text contents in an electronic Health Record (HER) systems) real time data which cannot be handled by standard database management systems like DBMS or RDBMS, an innovative database structure need be placed in order to streamline and eliminate redundancy, inaccuracy, and enable to have a single version of the truth of data. One of the fundamental issue in working with very large healthcare data, e.g. in the terabyte or petabyte range, small inefficiencies in storing data can have a large effect on ability to retrieve and process these data for other analysis. Third, in the knowledge/data process and logical capacity domain, the traditional operational focus needs to be shifted to a more analytic focus that could manipulate and convert various types of unstructured data and metadata into information context and actionable knowledge [56,57].
Last, but not least, in the resources domain and from the culture perspective, an integrative level has to be reached and shifted from personal/individual level with organizational and systematic approach where data is viewed as an asset with analytical culture and high predictive value [59,60]. Note that above four level hierarchical infrastructures of Big Data science determines it as a connection and systematic science merging and integrating cutting edge diverse multidisciplinary fields for better informed and shared decisionmaking (Table 1 for more examples, cases, software and relevant references).
Hierarchical Domains
Software
Features/Tasks/Outcomes
Examples, Cases, Resources
Some References
Platforms,Hardware, Physical Capacity
Hadoop system
Parallel distributed, multicore, cloud and clustering for timeliness, privacy, transparency, data sharing, and integrity
Map-Reduce framework: Open flow application programming to interfaces or a virtual network overlay for controlling, understanding and dealing with Big Data, which could also create agility and automation with a centrally programmable network
[43-49]
Data Storage Databases
HDFS distributed file systems
Storage, backup, retrieval, acquisition, formatting to remove redundancy, inaccuracy
Big data Script Database: DBMS, RDBMS or ORDBMS
[50-53]
Fundamental Data Preprocessing
R/pbdR; bioconductor, SAS JMP, SPSS, Matlab
Data cleaning, extracting, integration, aggregation, visualizations
Software-defined data center or software-defined network, SoFIA, ExScalibur
[ 25,35,72-74,80-83]
Advanced Computational Approaches
R/pbdR; Revolution Analytics, Tableau Software, SAS JMP, Matlab
Modeling, analysis, computing, interpretations
Network and systematic based approaches
[13,75-79,84-87]
Resources Domain, Data/ Variable Types and Applications
Bioconductor/R; BRB-ArrayTools
Low cost, reduced medical error, actionable knowledge high predictive value
1. OMICS, Precision Medicine, The Cancer Genome Atlas(TCGA) project (large n, large p: next generation sequencing, genomes, transcriptomes, epigenomes from cells, tissues and organisms)
[1-5,24,88,89]
Heterogeneity
2. Comparative Effectiveness and Patient Centered OutcomeResearch, implementation sciences(large n, large p): hospital, lab, biometric data such as finger prints, handwriting, retinal scans, X-ray and other medical images, pulse-oximetry readings, and other unstructured, semi structured, health device, media or censored and EHR data
[4-6,11,14,18-20]
Three Vs, large n, large p
3. Human genomics: clinical trial or animal study (large p, small n)
[52,75-82,84-87]
Table 1: Table 1: Big Data Domains, Features, Software/Hardware, Analytical Approaches, and Examples/Applications.
Big data science debates, challenges, and opportunities
Big Data science is now considered as “interdisciplinary fields work principally in the social sciences, humanities and computing and their intersections with the natural sciences about the implications of Big Data for societies” [26]. Due to its real time nature, and rich information enabled by new technologies, Big Data science has potential to offer a higher form of intelligence and knowledge with the aura of truth, objectivity, and accuracy [61,62]. Currently, there is a good understanding that addressing researcher’s subjectivity with Big Data sciences could make research more scientific, robust, and ethical. However, how real time features shaping the researchers’ usage of Big Data during gathering, manipulating, analyzing, and visualization process could be a challenging issue, and need to be examined.
External factors or data types, e.g., in the social media contents for the health related issues, the streaming unstructured user-generated text based qualitative data derived from subjective perceptions and personal experience may interfere and paint data with a misleading picture, and, in the end, what it quantifies does not necessarily have a closer claim on objective truth. Therefore, developing conceptual models grounded in the complex and unstructured data in the qualitative research perspective for detecting the subjectivity, the external factors, and abnormality of Big Data that may affect outcomes is really in need, and might be new research opportunities [35].
Moreover, since Big Data is not a random sample, but contains all data, ‘The Age of Big Data’ explosion raises some debates and challenges regarding the need of new scientific computational methods, and the values of the traditional statistical inference theories that has prevailed for centuries in data sciences, but now might be outdated [63-66]. We all know that the Big Data era requires exhaustive, to the plenary, unlike the random sampling based traditional statistical approaches. Should the best analytical approach in this new big data era be exhaustive using of full data with more intelligent (be specific, artificial intelligence or machine learning based) rather than random sampling the big data?
To answer why plenary exhaustive might be more valuable, we may take a look at an evidence-based practice/medicine example. Based on the BMJ online forum, seventy five percent of doctors believe that adverse consequences has led the evidence-based practice/medicine moving toward collapse, and one real challenge is not evidence-based medical system itself, but that it is being improperly used due to the fact that most patients do not meet the clinical study inclusion criteria and most real cases are being considered as outliers. It is known that statistical significance does not imply the clinical significance, and correlation doesn’t conclude causal relationship.
Note that a common ending for either Big Data or traditional sampling based inference in medical science is that 1) as the sample/ data size grows larger, the science gets stronger; 2) follow-up time (real) the longer, the results are closer to clinical, and the greater value for clinical significance and usefulness.
Therefore, as an important inevitable complementary, Big Data science may overcome some challenges in evidence-based medical system (practice or medicine), and should be emphasized from research and clinical perspective with better data sharing and security plan, transparency, and integrity. This is because not only Big Data science allows researchers to study treatment effectiveness, and patient heterogeneity, but also the need for treatments to be allocated by randomization with continuously arriving new sample. In addition, through the integration of large data from published literatures and meta-analysis, secondary literature conclusions reached as a use of scientific methods to guide clinical practice itself could have important clinical significance and scientific value.
On the other hand, traditional statistical inference perspective, an important merit that Big Data science brings in is that it allows continuous refinement of the computational or statistical model and the associated assumptions with continuous arrival of new data for more accurate outcome and better informed decision making due to its real time, evolving and dynamic feature. More importantly, it allows applying predictive analytics to understand not only what has happened and what is currently happening, but also to predict what will happen in the future. The key challenges researchers face today in the area of Big Data is still the ability of researchers to locate, analyze, integrate, and interact with all real time data and associated software due to the lack of adaptive intelligent tools, accessibility, and appropriate training at the current stages [67,68].
In order to overcome such challenge for interpretable outcomes and replicable or reproducible results, and arriving to actionable and accurate medical decision making, close multidisciplinary collaborations of Big Data analysts with domain experts are needed. First, traditional data analysts (e.g., statisticians and mathematicians) should join with the new evolving class of “data scientists” (e.g., computer scientist/engineers) and create intelligent automatic systems and high level adaptive analysis tools to make full use of the Big Data and let the data speak for itself. Second, the domain experts including biomedical, social/behavioral scientists and scientists in economics, business, and geosciences, etc. need to work closely with Big Data scientists to make sense of the big data in order to extract actionable knowledge. The next generation of good Big Data scientists are indeed in demand of persons with brains for math, skills with computers, eyes of artists and abilities to: i) write algorithms that filter data; ii) churn through billions or trillions of data points and show where patterns emerge and what matters; iii) understand what they are telling; iv) graphically represent the information; v) make the judgment more sound, and more objective that may lead to better decision-making [69].
Hospitals throughout the United States currently undergo major operational change in order to complete Electron Health Record implementations and demonstration of their Meaningful Use in order to qualify for Centers for Medicare & Medicaid Services Incentive Programs and to avoid penalties [58]. Hospital administrators typically do not have additional resources to perform their own Big Data Analysis and is not part of their scope of work [59]. Also, due to the variety of the EHRs which are being used at the different hospitals and the current lack of Health Information Exchange among vendors and EHR products, Big Data Analysis of hospital multisite EHR and other data are rare and difficult to perform. Even single site Big Data Analysis is often done by researchers or employees who would like to answer specific questions, as opposed to being done by the vendors or by the hospital administrators [59].
However, if and when Health Information Exchange finally happens, the doors will suddenly open to Big Data Analysis that is expected to have huge positive implications to knowledge generation that shall impact research and practice. Some of the current problems are that in the past and in the present, hospital data was guarded due to HIPAA, conflict of interest, and its potential negative financial implications to the owner institutions. Vendors also have motivation to not develop EHR and other software systems that are interoperable with other software systems developed by other vendors, since that would make it easier for hospitals to change vendor in the future, which would have negative financial impact on the vendor [60].
Big data analytic approaches
Ultimately, the value of Big Data is not about the Big Data, it’s about how to turn big data into good research problems/questions/ hypotheses, then transform into valuable solutions that benefit society [70,71]. This is rendered simpler by their applications, for instance, the rapid advance of EHRs, mHealth, eHealth, Smart and Connected Health, and telehealth devices merging with social, behavior science, genomics and economics have led to the development of new infrastructure and transformation of health care systems for precision medicine and better-individualized patient care.
One important question for Big Data scientists to ask: 1) How to transform some 300 billion data points into quantitative statistical evidence for diagnostics, therapeutics, and new insights into population health, disease and treatment? 2) What are the best approaches? Does the traditionally used inference technique continue to play some roles? For instance, should it be experimental versus computational; hypothesis driven versus data driven; traditional statistical modeling versus data mining and artificial intelligence approaches.
To make the overwhelming volume of Big Data actionable and analytics operational, several key issues of how we proceed and analyze the data requires special attentions. First, bottleneck of the Big Data: Analysis tools and the development of advanced statistical and computational techniques with pipelines that can easily scale up with the three V’s (Volume, Velocity, Variety) and its complexity. These tools make high-powered methods available to not only professional statisticians, but also to casual users. Second, creator of Big Data value is the integration and linkage of heterogeneous Big Data, which has formidable logistical and analytical challenges. Third, validation, interpretation, and visualization: are crucial to extracting actionable knowledge for decision making which require Big Data analysts to closely collaborate with domain experts.
Therefore, in order to transform the billions of data points into valuable and actionable solutions require deeper learning and data analysis at both fundamental and advanced levels [25,72-74]. The fundamental level analysis include 1) basic online real time queries, pipeline, flow, analysis tools; 2) data pre-processing or big data reduction: detecting the missing data, errors, outliers; extracting, transforming, loading part of data preprocessing, automated filtering of non-useful data, redundancy and correlations; 3) computational techniques for summarizing the qualitative and quantitative results, unveiling trends and patterns, and generating reports; 4) data automations and generations for metadata, e.g., computer-automated analysis of blog postings; 5) visualization tools with simple and easy models: interpreting and making sense of the data.
At the advanced level data analysis: systems based and network approaches for data integration in genomic research is a good example. The followings are lists but not limited potential sophisticated computational and statistical approaches 1) Real time analytics and Meta-analysis that integrates multiple data sources including bedside healthcare streaming data; 2) hierarchical or multi-level model for spatial (state and national) data; longitudinal and mixed model for real time or temporal dynamic data rather than static data; 3) data mining, pattern recognitions for trends, and pattern detection; 4) natural language processing for text data mining; machine learning, statistical learning, Bayesian learning with auto-extraction of data and variables; 5) artificial intelligence with deep learning (e.g., neural network, support vector machine, dynamic state space model), automatic ensemble techniques and intelligent agent for automated analysis and information retrieval; 6) causal inferences and Bayesian approach with probabilistic interpretations [13].
Comparing fundamental level analytic with advanced level analytic in Big Data science, fundamental analytic including descriptive analytics serves for the purpose to summarize “what has happened” (e.g., in a simplest type that allows you to break down big data into smaller, more useful pieces of information) and focus on the insight gained from historical data to provide trending information on past or current events (e.g., looks at data and information to describe the current situation in a way that trends, patterns, and exceptions become apparent). While the advanced level computational tools listed above in Big Data science focuses on predictive and prescriptive analytics, which intends to determine patterns and predict future outcomes and trends, and answers “what could happen” and “what should we do?” through quantifying effects of future decisions in order to advise on possible outcomes. Prescriptive Analytics includes functions as a decision support tool by exploring a set of possible actions and suggesting actions based on descriptive and predictive analyses of complex data. It also conducts real-time analytics by using point-of-care data and analyzes the data at the point of care to present immediate and actionable information to providers.
Human genomics/OMICS application and example
Patient centered Electronic Health Records (EHR) big data examples have been reviewed and discussed recently, mainly for the case of large sample size n in terms of three V’s, but not for large number of parameters p [2-6]. Therefore, here we focus on a human OMICS (large “p”: next generation high-throughput sequencing data, genomes, transcriptomes, epigenomes and other omics data from cells, tissues and organisms) in personalized and precision medicine research as an important application and great example of Big Data science in the medical fields. Figure 1 demonstrates this translational research scheme/process from Big data generating instrument/technology, to complex data preprocessing and analytical pipeline, procedures and approaches in order to transform 300 billion data points of disease data into diagnostics, therapeutics, and new insights into population health and disease treatment. We can see that both lower level fundamental analysis and high level advanced computational analytic tools could play significant roles and be a more cost effective way than experimental ways in the big data world.
Figure 1: Big Data science in the fields of biomedical research: Transforming big genome data into diagnostics, therapeutics, and new insights into population
health, disease treatment.
Prior to performing advanced analytic techniques, to guarantee the quality of the Big data, various preprocessing at fundamental data analysis that combines, pools, aggregates and transforms various raw data into appropriate data format need be done. For instance, in the genomic domain, for Affymetrix time course data obtained from Affymetrix GeneChips, one may use Affymetrix software (MAS 5.0) and probe set algorithms of MAS5 for background subtraction, signal intensity normalization between arrays, and non-specific hybridization correction etc [75-79]. To do so, high level performance hardware and software (e.g., programming languages and algorithms for visualizations) that conduct parallel and distributed and cloud computing to manage, retrieve, reformat and analyze the data from various resources including the genomic laboratory and hospital patient information systems needs be considered (Table 1) [58,59,80- 84]. For instance, Bianchi et al. [81] developed HTS-flow, a workflow management system that can retrieve information from a laboratory management system database, manages Omics data analyses through a simple GUI, outputs data in standard locations and allows the complete traceability of datasets, accompanying metadata and analysis scripts. Childs et al. [82] designed and implemented SoFIA, an Omics data integration framework for annotating high throughput data sets [82]. Kovatch et al. also shared their experiences designing an optimized whole genome DNA and RNA pipeline system for the “Genome Analysis ToolKit (GATK) Best Practices” and provided an evaluation of computing workload and I/O characteristics [83].
Besides the above discussed fundamental analysis,from thousands of genes to identify a handful of genes responded to the drug over time that could be potential drug targets could turn into a computational problem related to the “curse of dimensionality” issue (large “p”) in the temporal fashion. Various statistical/machine learning and data mining techniques or statistical testing approaches could be applied and compared for addressing such to examine the reproducibility issues including: 1) Data driven (mining) versus hypothesis driven (testing); 2) unsupervised learning (clustering) versus supervised (classifications); 3) optimization versus sequential or recursive feature reduction with multiple testing: i) linear versus nonlinear model; ii) parametric, nonparametric, semi-parametric statistical model with L-norm regularization techniques; iii) univariate versus multivariate methods; iv) Bayesian with prior knowledge/distribution versus non- Bayesian/classical statistical approaches; v) Hierarchical Bayesian with shrinkage in statistical modeling versus Automatic Relevance Determination in neural network.
Here we briefly present a simplified example of “large p” through comparisons of various statistical methods for multiple sclerosis disease studies in human genomics [84]. The genome data set contained gene expression data from 14 MS patients given a 30g dose of intra-muscular IFN1a and the gene expression data available for 10 time points: before treatment, 1h (hour), 2h, 4h, 8h, 24h, 48h, 5d, 7d & 3months. After data preprocessing and filtering from millions gene, 4324 genes measured at 10 time points on 14 patients with a total of 605,360 measures or data points were included for further data analysis. The key biological questions of this study are 1) the identifications of significant differentially expressed genes responding to the treatment, and 2) characterizing the dynamics and changes of gene expression to determine the trajectories of significantly regulated genes in responding to the treatment.
For comparison purposes, we presented the following six computational methods for the “curse of dimensionality” issue in the temporal fashion in order to identify a handful of genes responded to the drug over time from thousands of measures: 1) parametric methods with the Analysis of Variance (ANOVA) with bootstrapping resampling techniques; 2) semi-parametric with class dispersion method; 3) nonparametric with Pareto with permutation methods; 4) mixed effects model (non-Bayesian) with bootstrap; 5) Bayesian linear correlated/multivariate model; 6) Bayesian nonlinear model. Figure 2 provides the condensed results of each method to demonstrate their differences, note that all are adequate in capturing and identifying the significant/relevant genes responding to the treatment and disease progression.
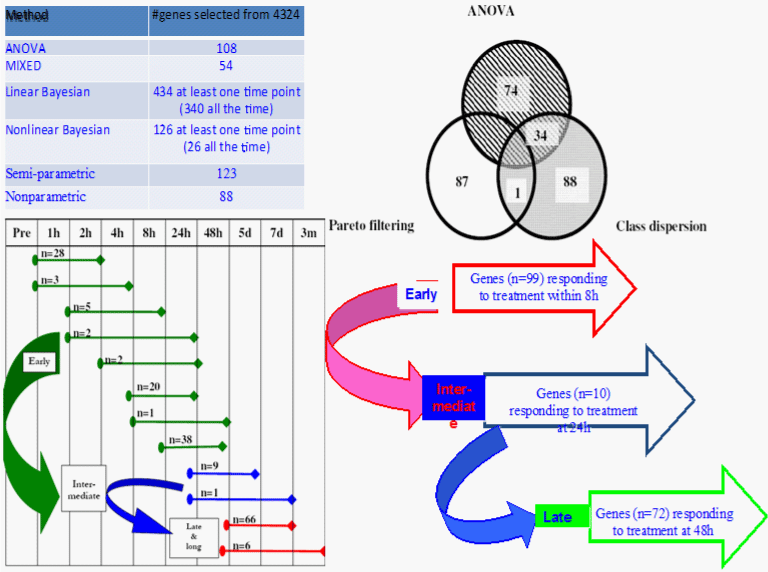
Figure 2: Comparison of gene selection/filtering methods in time course gene expression data: Identifications of significant differentially expressed genes in
response to the treatment from various methods (top); post clustering and path analysis for characterizing the dynamics and changes of gene expression from early
stage to intermediate to the late stage to determine the trajectories of significantly regulated genes to better understand the treatment effect (bottom).
For the parametric method: mixed models proved to be more conservative. For the semi-parametric with class dispersion and nonparametric with Pareto methods are appropriate in capturing variation from time to time, thereby making them more suitable for investigating significant monotonic changes and trajectories of dynamic changes. Simulation studies showed that the semiparametric with class dispersion performs best regarding robustness of rejection of hypothesis given different significance (alpha) levels, while parametric ANOVA and nonparametric Pareto perform similar. For nonlinear Bayesian versus linear Bayesian multivariate model is more conservative but more robust, and perform better with regard to different type I error rates while linear model showed better goodness of fit than nonlinear model.
Moreover, post clustering and path analysis is able to not only identify the genes that are over expressed, under-or not expressed, but to isolate trajectories of genes whose regulations appear to be interdependent, inferring the possible inter-gene-dependence pathway and network showing early, intermediate, and late gene clusters to better understand the treatment effect. In short, the combinations of these various approaches would provide us more comprehensive picture of the solutions and reliable results that illustrates the values and roles of the advanced computational tools transforming thousands of Big Data points into quantitative statistical evidence for diagnostics, therapeutics, and new insights into disease, population health, and treatment [75-79,85-87]. Health/nursing and medical researchers could employ these advanced analytical tools in Big genome research for either disease specific (e.g., neurology conditions, cancer, cardiovascular diseases) or domain specific such as pain, fatigue, physical functioning or multiple chronic health conditions.
Conclusion
Big Data has the potential to impact various fields from social science to political science, from financial industry to business, from medical science to public health, from health care to genetics, and from personalized medicine to patient/custom-centered outcomes. It has involved various levels of human life: individuals to community, and industrial to university to government. The emerging field of Big Data science and associated practices offered new opportunities and is promising, but it comes with many challenges in all fields, especially the biomedical and health science fields which makes improved understanding of human life, health, diseases, and behavior possible. The collaborative network, nurturing environments and interdisciplinary, team-science approach with highly trained computational skills and domain/disease expert talents are crucial, while adaptive and intelligent evolving analytic tools and smart utilization of open resources are keys for enhancing the true value of real time big data for actionable healthcare decision making and better informed patient outcomes.
References
- Brabham DC, Ribisl KM, Kirchner TR, Bernhardt JM. Crowd sourcing applications for public health. American Journal of Preventative Medicine. 2014; 46: 179-187.
- Lissovoy G. Big data meets the electronic medical record: A commentary on identifying patients at increased risk for unplanned readmission. Medical Care. 2013; 51: 759-760.
- Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inf Sci Syst. 2014; 2: 3.
- Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA. 2013; 309: 1351-1352.
- Bates D, Saria S, Ohno-Machado L, Shah A, Escobar G. Big data in health care: Using analytics to identify and manage high-risk and high-cost patients. Health Affairs. 2014; 33: 1123-1131.
- Gardner E. The HIT approach to big data: Everyone in this market is trying to corral their massive data sets. Health Data Management. 2013; 21: 34.
- Laney D. 3D Data Management: Controlling Data Volume, Velocity and Variety. Gartner. 2001.
- Laney D, Beyer MA. The Importance of 'Big Data': A Definition. Gartner. 2012.
- Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003; 422: 835-847.
- McElheny VK. Drawing the Map of Life: Inside the Human Genome Project. Basic Books. 2010.
- Snyder M, Du J, Gerstein M. Personal genome sequencing: current approaches and challenges. Genes Dev. 2010; 24: 423-431.
- Lohr S. The origins of ‘big data’: An etymological detective story, New York Times. 2014.
- Bennett CC, Hauser K. Artificial intelligence framework for simulating clinical decision-making: a Markov decision process approach. Artif Intell Med. 2013; 57: 9-19.
- Hampton T. Disease, drug response linked to loss or gain of big DNA chunks in genome. JAMA. 2007; 297: 1539-1540.
- Bachrach Y, Kosinski M, Graepel T, Kohli P, Stillwell D. Personality and patterns of Facebook usage, WebSci ’12: Proceedings of the 3rd Annual ACM Web Science Conference. 2012; 24-32.
- Bollen J, Mao H, Zeng X-J. Twitter mood predicts the stock market, Journal of Computational Science. 2011; 2: 1-8.
- Kuehn BM. NIH Recruits Centers to Lead Effort to Leverage “Big Data” JAMA. 2013; 310: 787.
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015; 372: 793-795.
- Marx V. Biology: The big challenges of big data. Nature. 2013; 498: 255-260.
- Weber GM, Mandl KD, Kohane IS. Finding the missing link for big biomedical data. JAMA. 2014; 311: 2479-2480.
- Daughtery SE, Whaba S, Fleurence R. Patient-powered research networks: building capacity for conducting patient-centered clinical outcomes research. JAMA. 2014; 21: 583-586.
- Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014; 21: 578-582.
- Clancy C, Collins FS. Patient-Centered Outcomes Research Institute: the intersection of science and health care. Sci Transl Med. 2010; 2: 37cm18.
- Schneeweiss S. Learning from big health care data. The New England Journal of Medicine. 2014; 370: 2161-2163.
- Hilbert M. Big Data for Development: From Information-to Knowledge Societies", SSRN. Rochester, NY: Social Science Research Network. 2013.
- Ruppert E. Big Data & Society. University of London, UK, SAGE, publisher. 2014.
- Jain SH, Rosenblatt M, Duke J. Is big data the new frontier for academic-industry collaboration? JAMA. 2014; 311: 2171-2172.
- Kvochko E. Four Ways to talk about Big Data (Information Communication Technologies for Development Series). 2012.
- Ohm P. Don't Build a Database of Ruin. 2012.
- Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, et al. Big data: The future of biocuration. 2008; 455: 47-50.
- Helles R, Jensen KB. Introduction to the special issue: ‘Making data-Big data and beyond’, First Monday. 2013; 18.
- Jacobs A. The Pathologies of Big Data. 2009; 7: 10.
- National Science Foundation (NSF) NSF Leads Federal Efforts in Big Data. 2012
- Wills MJ. Decisions through data: Analytics in healthcare. Journal of Healthcare Management. 2014; 59: 254-262.
- UN GLobal Pulse, Big Data for Development: Opportunities and Challenges (White P by Letouze E). New York: United Nations. 2012.
- Magoulas R, Lorica B. Introduction to Big Data. Release 2.0 (Sebastopol CA: O’Reilly Media). 2009; 11.
- Boyd D. Privacy and Publicity in the Context of Big Data. 2010.
- Beyer M. Gartner Says Solving 'Big Data' Challenge Involves More than Just Managing Volumes of Data. 2011.
- Snijders C, Matzat U, Reips UD. ‘Big Data’: Big gaps of knowledge in the field of Internet. International Journal of Internet Science. 2007; 7: 1-5.
- Bryant A, Charmaz K. Introduction: Grounded theory research: Methods and Practices. In: Antony Bryant, Kathy Charmaz. 2007.
- Bryant A. Re-grounding grounded theory, Journal of Information Technology Theory and Application. 2002; 4: 25-42.
- Boyd D, Crawford K. Critical Questions for Big Data. Information, Communication & Society. 2012; 15: 662.
- Hellerstein J. Parallel Programming in the Age of Big Data. Gigaom Blog. 2008.
- Assuncao D, Calheiros RN, Bianchi S, Netto MAS, Buyya R. Big Data Computing and Clouds: Challenges, Solutions, and Future Directions. Marcos Technical Report CLOUDS TR2013-1, Cloud Computing and Distributed Systems Laboratory, The University of Melbourne. 2013.
- O'Driscoll A, Daugelaite J, Sleator RD. 'Big data', Hadoop and cloud computing in genomics. Journal of Biomedical Informatics. 2013; 46: 774-781.
- Hashem IAT, Yaqoob I, Anuar NB, Mokhtar S, Gani A, Khan SU. The rise of “big data” on cloud computing: review and open research issues. Information Systems. 2015; 47: 98-115.
- Boja C, Pocovnicu A, Batagan L. Distributed Parallel Architecture for Big Data, Informatica Economica. 2012; 16: 116-127.
- Nielsen L. Hadoop: The Engine That Drives Big Data. Kindle Edition. 2013a.
- Nielsen L. The Little Book of Cloud Computing, Including Coverage of Big Data Tools. Kindle Edition. 2013b.
- Volk T. The Software-Defined Datacenter: Part 2 of 4 - Core Components. 2013.
- Cherian B. What Is the Software Defined Data Center and Why Is It Important? 2014; All Things D post. 2013.
- Cingolani P, Sladek R, Blanchette M. Big Data Script: a scripting language for data pipelines. Bioinformatics. 2015; 31: 10-16.
- Eddelbuettel D. High-Performance and Parallel Computing with R. CRAN Task View: High-Performance and Parallel Computing with R.
- Raim AM. Introduction to distributed computing with pbdR at the UMBC High Performance Computing Facility. Technical Report HPCF-2013-2. 2013.
- Markham AN. Undermining ‘data’: A critical examination of a core tem of scientific inquiry, First Monday. 2013; 18: 10.
- MIKE2.0. Big Data Definition. 2014a.
- MIKE2.0. Big Data Solution Offering. 2014.
- Health IT.gov. Meaningful Use definition & objectives. 2016.
- Moerbe M, Kelemen A. Turning electronic health record data into meaningful information using SQL and nursing informatics. Comput Inform Nurs. 2014; 32: 366-377.
- Trotter F, Uhlman D. Interoperability. Oram A. In: Meaningful Use and beyond: A guide for IT staff in health care. 2011; 165-200.
- Boellstorff T. Making big data, in theory. The First Monday. 2013; 18: 10.
- Hey T, Tansley S, Tolle K. The Fourth Paradigm: Data-Intensive Scientific Discovery. 2009.
- Anderson C. The long tail, Wired. 2004; 12: 10.
- Anderson C. The end of theory: The data deluge makes the scientific method obsolete, Wired. 2008; 16: 106-129.
- Steadman I. Big data and the death of the theorist. 2013.
- Graham M. Big data and the end of theory? 2012.
- Manyika J, Chui M, Bughin J, Brown B, Dobbs R, Roxburgh C. Big Data: The next frontier for innovation, competition, and productivity. 2011.
- WEF (World Economic Forum) & Vital Wave Consulting. Big Data, Big Impact: New Possibilities for International Development. 2012.
- Nielsen L. Unicorns among Us: Understanding The High Priests of Data Science. 2014.
- Asay M. Data scientists: Do they even exist? Data everywhere, but not a drop to shrink. 2013.
- Asay M. Big data myths give way to reality. 2013.
- Martinez MG, Walton B. The wisdom of crowds: The potential of online communities as a tool for data analysis. Technovation. 2014; 34: 203-214
- Billings SA. Nonlinear System Identification: NARMAX Methods in the Time, Frequency, and Spatio-Temporal Domains. 2013; 574.
- Vis F. A critical reflection on Big Data: Considering APIs, researchers and tools as data makers. 2013; 18: 10.
- Liang Y, Kelemen A. Bayesian state space models for inferring and predicting temporal gene expression profiles. Biom J. 2007; 49: 801-814.
- Liang Y, Kelemen A. Bayesian Models and Meta Analysis for Multiple Tissue Polygenic Gene Expression Data Following Corticosteriod Administration. BMC Bioinformatics. 2008; 9: 354.
- Liang Y, Kelemen A. Statistical Advances and Challenges for Analyzing Correlated High Dimensional SNP Data in Genomic Study for Complex Diseases. Statistics Surveys. 2008; 2: 43-60.
- Liang Y, Kelemen A. Bayesian finite Markov mixture model for temporal multi-tissue polygenic patterns. Biom J. 2009; 51: 56-69.
- Liang Y, Kelemen A. Sequential Support Vector Regression with Embedded Entropy for SNP Selection and Disease Classification. Stat Anal Data Min. 2011; 4: 301-312.
- Bao R, Hernandez K, Huang L, Kang W, Bartom E, Onel K, et al. ExScalibur: A High-Performance Cloud-Enabled Suite for Whole Exome Germline and Somatic Mutation Identification. PLoS One. 2015; 10: e0135800.
- Bianchi V, Ceol A, Ogier AG, de Pretis S, Galeota E, Kishore K, et al. Integrated Systems for NGS Data Management and Analysis: Open Issues and Available Solutions. Front Genet. 2016; 7: 75.
- Childs LH, Mamlouk S, Brandt J, Sers C, Leser U. SoFIA: A Data Integration Framework for Annotating High-Throughput Data Sets. Bioinformatics. 2016.
- Kovatch P, Costa A, Giles Z, Fluder E, Cho HM, Mazurkova S. Big omics data experience Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis. 2015.
- Liang Y, Tayo B, Cai X, Kelemen A. Differential and trajectory methods for time course gene expression data. Bioinformatics. 2005; 21: 3009-3016.
- Liang Y, Kelemen A, Tayo B. Model Based or Algorithms Based? Gene Expression Based Statistical Methods to Find Evidence of Diabetes. Journal of Statistical Methods for Medical Research. 2007; 16: 139-153.
- Kelemen A, Liang Y, Vasilakos A. Review of Computational Intelligence for Gene-Gene Interactions in Disease Mapping, in “Computational Intelligence in Medical Informatics”. Kelemen A, Abraham A, Chen Y, Editors. In the Series in Studies in Computational Intelligence. 2008; 1-16.
- Kelemen A, Vasilakos A, Liang Y. Computational Intelligence for Genetic Association Study in Complex Disease: Review of Theory and Applications. International Journal of Computational Intelligence in Bioinformatics and Systems Biology. 2009; 1: 20-36
- Zhao Y, Simon R. BRB-ArrayTools Data Archive for human cancer gene expression: a unique and efficient data sharing resource. Cancer Inform. 2008; 6: 9-15.
- Cancer Genome Atlas Research Network. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015; 163: 1011-1025.