
Mini Review
Austin Biomol Open Access. 2016; 1(1): 1004.
Using Metal Nanoparticles to Interrogate Transcription Factors - DNA Interactions for Cancer Research – A Mini Review
Xiaodi Su1,2*
¹Institute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, #08-03 Innovis, Singapore 138634
²Department of Chemistry, National University of Singapore, Block S8, Level 33 Science Drive 3, Singapore 117543
*Corresponding author: Xiaodi Su, Institute of Materials Research and Engineering, Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, #08-03, Innovis, Singapore
Received: August 14, 2016; Accepted: October 25, 2016; Published: November 01, 2016
Abstract
Transcription factors are DNA binding proteins, responsible for gene transcription. In this mini review, four gold nanoparticle (AuNPs)-based bioassays developed in our group are described. They can collectively answer the following questions for transcription factor-DNA interactions, (1) sequence specificity, (2) ligand effect, (3) sequence rule down to single base resolution and (4) co-operative and/or site specific protein bindings to composite DNA. These assays use either AuNPs aggregation induced color change (colorimetric assays) or AuNPs supported Nanomaterial Surface Energy Transfer (NSET)/ Forster Resonance Energy Transfer (FRET) principle. These assays are compared in their detection principle, usage (what characteristics can be deducted), strengths and limitations. Similar work from other groups will be also included to paint a full picture as for the powerful utility of metal nanoparticle in interrogating protein-DNA interactions. With this review one can gain an insight of how metal nanoparticles’ optical properties can be harnessed to design versatile bioassays (Table of Content Abstract).
Keywords: Gold nanoparticles; Protein; DNA; Transcription factors; Cofactors
Abbreviations
AuNPs: Gold Nanoparticles; AgNPs: Silver Nanoparticles; mNPs: Metal Nanoparticles; ERa : Estrogen Receptor a; ERΒ: Estrogen Receptor Β; ERE: Estrogen Response Elements; TF: Transcription Factor; NSET: Nanomaterial Surface Energy Transfer; FRET: Förster Resonance Energy Transfer; ELISA: Enzyme-Linked Immunosorbent Assay; FoxA1: Forkhead Box A1; AP2γ: Activating Protein 2γ; FID: Fluorescent Intercalator Displacement; TO: Thiazole Orange
Introduction
Transcription factors are DNA binding proteins, responsible for gene transcription. They are key factors in many cellular processes such as growth and cell development, intra and extracellular signaling and cell cycle. Several diseases have been linked to transcription factors malfunction (e.g. cancer, congenital heart disease, and renal malfunction etc). In a classical mode, TFs regulate gene transcription by binding to their specific DNA elements in many other modes, gene transcription is regulated by TFs and co-factors by cooperative interactions with DNA [1]. Figure 1 is a schematic example of the actions related to estrogen receptors (ERa and ERΒ), ligand activated nuclear hormone receptors. In the classical mode of estrogen gene regulation ER binds to its DNA element containing a core sequence of palindromic repeats separated by a three-base spacer (5’-GGTCAnnnTGACC-3’). The key questions are (1) the sequence rule and/or how base alternation in the core sequence will affect the binding affinity and (2) how ligand loading to ERs will affect their interactions with DNA. In the non-classical mode, key research is to identify co-factors and understand the cooperative binding between multiple proteins. Site selectivity and optimal distance between two sites for the best collaborative binding of multiple proteins are important in revealing gene regulation principle and for develop novel therapeutics. In general, a comprehensive understanding of TFs-related molecular binding events and their impact in gene transcription is beneficial to cancer research. For example, identifying the sequence rule of TFs-DNA binding would allow one to determine primary genes that govern cell division and are responsible for cancer [2]. Studying drug loading effect on TF-DNA interaction would lead to the understanding of how cancer drug exert effect. Identification of co-factors that collaboratively bind to DNA site would lead to the development of new therapeutic drugs targeting the co-factors [3].
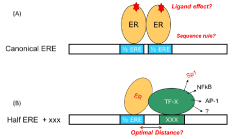
Figure 1: (A) Classical and (B) non classical mode of hormone gene regulation
by estrogen receptor. The canonical ERE contains palindromic repeats (half
ERE or ½ ERE) separated by a three-base spacer (5’-GGTCAnnnTGACC-3’).
Conventional biochemistry methods, e.g. Electrophoretic Mobility Shift Assay (EMSA), gel filtration chromatography assay and ELISA etc. have been extensively used in biological research labs to determine protein-DNA interactions. Biophysical techniques, Surface Plasmon Resonance Spectroscopy (SPR), Dual Polarization Interferometry (DPI) and Quartz Crystal Microbalance (QCM) are also capable of real-time measurement of these affinity binding events [4-6]. However, the tedious procedures (labeling, staining, surface modification, multiple incubation/washing steps, etc) and the requirement of sophisticated laboratory settings and equipment have made these technologies less appealing for high throughput and convenient usage in less well equipped laboratories.
Nanomaterials and nanotechnologies have led to a great deal of technology advancement in many fields, including analytical and bioanalytical sciences. Nanomaterials unique physical properties (optical, electrical, magnetic etc) have been largely exploited for biosensor and bioassay development [7-9]. Gold nanoparticles (AuNPs) have unique optical properties arising from their ability to support Localized Surface Plasmon Resonance (LSPR). AuNPs can be used as colorimetric sensor probe to detect bioaffinity interactions (i.e. DNA-DNA, DNA-metal ions, DNA-DNA binding drugs, protein-DNA, peptide-sugar, and bacteria etc) and monitor biological processes (i.e. protein phosphorylation, DNA cleavage etc), exploiting interparticle distance-determined solution color change. AuNPs can also support fluorimetric assays due to their super quenching to approximate fluorophores.
In the past few years, we have developed a suite of AuNPs-based assays for study bioaffinity interactions [10-23]. Working extensively with ERs and related co-factors, we have demonstrated versatile assay designs that can answer many questions, as represented in Figure 1, without using sophisticated equipment and yet in a rapid and quantitative manner. In this mini review we will describe four AuNPs-based bioassays that can collectively answer the following questions (1) sequence specificity in protein-DNA interactions, (2) ligand effect on protein-DNA binding, (3) sequence rule down to single base resolution, (4) cooperative and/or site specific ER and cofactor bindings to composite DNA. These assays use either AuNPs aggregation induced color change (colorimetric assays) or AuNPs supported Nanomaterial Surface Energy Transfer (NSET)/ Förster Resonance Energy Transfer (FRET) principle. We have compared the four assays in their detection principle, usage (what characteristics can be deducted), strengths and limitations. Similar work from other groups will be also included to paint a full picture as for the powerful utility of metal nanoparticle in bioanalytical sciences, related to protein-DNA interactions.
Versatile AuNPs-based Bioassays
Screening protein-DNA interactions using unmodified AuNPs (Assay I) [10-12]
In this assay citrate ion-coated AuNPs (13 nm) was used as sensing material. The assay is based on our discovery that protein- DNA complexes can stabilize citrate ion-capped AuNPs against salt-induced aggregation to a larger extent than the protein or the DNA alone (Figure 2), due to their unique molecular size and charge properties that provides a strong electrosteric protection to the AuNPs. We have firstly developed the concept using ERa and ERΒ [10] and later further validated the assay for its generality using FoxA1 (Forkhead Box A1) and AP2γ (Activating Protein 2γ) [11]. With this assay, important parameters of protein-DNA binding events e.g. sequence selectivity, distinct DNA binding properties of protein subtypes, binding stoichiometry, and sequence-independent transient binding can be determined instantly without using labels, tedious sample preparations and sophisticated instrumentation. One key hypostasis of this assay principle is that protein-DNA complexes can easily absorb on AuNPs to provide the AuNPs with electro-static protection. This hypostasis has been affirmed through comprehensive characterization using DLS and zeta potential measurement of the particle size and surface charge in the presence of protein, dsDNA and protein-dsNDA complexes, separately [10,11]. Using organic dye labeled dsDNA to form protein and then measure the fluorescent emission when AuNPs is added, an obvious fluorescent quenching is observed, which confirm the attachment of the protein-dsDNA complex on the AuNPs [12] that can provide enhanced negative charge on AuNPs (measured by the zeta potential), which is not possible for dsDNA along (it is well know that dsDNA itself has limited affinity to AuNPs [22,23].
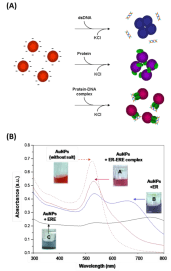
Figure 2: (A) Illustration of the principle of this bare AuNPs-based colorimetric
sensor for protein-dsDNA binding study. (B) UV-vis spectra of AuNPs mixed
with dsDNA, protein, or protein-dsDNA complex in the presence of NaCl of
100 mM [10] (reproduced with permission from ref. 8, @ 2010 American
Chemical Society).
This assay is easy to construct (using unmodified AuNPs) and is suitable for fast screening of DNA binding sequences to a purified protein in buffer solution. This assay has also been used to measure cooperative binding of two proteins to a composite DNA, exploiting distinct molecular size property of complexes with two proteins relative to one protein [12]. The binding stoichiometry is determined based on protein: DNA ratio, at which the protection effect is maximized, indicating that for the fixed amount of DNA protein binding is saturated at that ratio. The limitation of this method is however the use of unmodified AuNPs, making this method effective only for purified individual protein but not for nuclear extract.
Sensing transcription factor through controlled-assembly of metal nanoparticles modified with segmented dna elements (Assay II) [14,15]
Differing from the Assay I, where unmodified AuNPs is used, here this assay involves two sets of double-stranded (ds) DNA modifiedmNPs, each carrying a half site segment of the ER binding DNA. Each of these half sites is designed to contain a short complementary sticky end that introduces base-pairing forces to facilitate particle aggregation and to form a transient full dsDNA sequence. The detection of specific ER-DNA binding is founded on the premise that the mixture of these two sets of dsDNA-mNPs experiences a remarkable particle aggregation under certain salt conditions; whereas the aggregation can be retarded in the presence of specific protein that binds and stabilizes the transient full dsDNA structure and therefore introduces steric protection forces between particles. We have demonstrated the concept using both ERa and ERΒ with gold and silver NPs as the sensing platform. UV-vis spectroscopy, TEM and Dynamic Light Scattering (DLS) measurements were conducted to provide full characterization of the particle aggregation/dispersion mechanism. Differing from most of the mNP-based colorimetric sensors that are designed based on the analyte-induced aggregation mechanism, current protein binding-stabilization sensing strategy reduces the false signals caused by unrelated particle de-stabilizing effects.
To better understand the aggregation mechanism and its DNA design dependency we systematically investigated the effects of DNA conformation, number of sticky-ends bases, spacer length as well as their symmetrical and asymmetrical interplay between the complementary set of segmented DNA–AuNPs conjugates on particle aggregation kinetics [15]. A few conclusions are reached namely (1) dsDNA serves as a more effective spacer than ssDNA in preventing base coordination to the AuNPs surface, due to its rigidity that in turn helps to improve the accessibility of sticky ends for faster aggregation; (2) base-pairing force in facilitating the saltinduced AuNPs aggregation is tunable by the number of stickyends; (3) symmetrically spaced sticky-ends enable quicker noncrosslinking aggregation than the asymmetrical combination due to their closer initial interparticle distance. These conclusions are of significant for biomolecular interactions induced nanoassembly. This design concept has been further modified by using stoichiometric DNA-AuNPs conjugate (one DNA fragment on one AuNPs). Two sets of such conjugate can detect TFs binding using Dynamic Light Scattering (DLS), according to protein binding induced size change [16].
A hybrid sensor using dna-coated gold nanoparticles and water soluble conjugated polymers for studying proteindna interaction and ligand inhibition (Assay III) [17-19]
Both Assays I and II above are using metal NPs’ aggregation and inherent color change as measures of protein-DNA complex formation and protein-DNA binding. In this assay here, AuNPs’ fluorescent quenching is used to form a hybrid sensor in collaboration with water soluble Conjugated Polyelectrolytes (CPEs). It is based on the change of CPEs’ distance to dsDNA-conjugated AuNPs, upon protein binding to the dsDNA on AuNPs surface. A range of CPEs bearing with different either positive or negative charges, emitting at different wavelengths either far or close to AuNPs’ absorption wavelength 520 nm) are used. According to the initial interaction of the CPEs with the dsDNA-AuNPs, “light-on”, “light-off”, and “two way” assays have been constructed for proteins with known or unknown charge properties. The assays have been extended to screen small molecular weight ligands that can inhibit protein-DNA interaction, which is important in drug development. Figure 3 is an example of using a cationic CPE, of which the original emission is largely quenched due to close distance to AuNPs through the strong electrostatic interaction with the negatively charged DNA coating on gold. Protein binding (ER, AP2γ and FoxA1) to respective DNA is detected as increased fluorescent emission, i.e. CPE become further away from the AuNPs due to the reduced overall surface charge proteins are positively charged at the pH of the binding buffer.
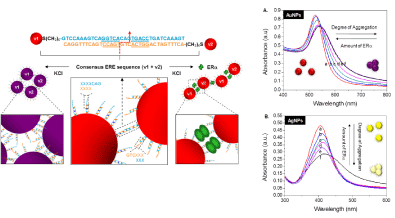
Figure 3: Schematic illustration of the sensing principle of assay using the segmented DNA-functionalized AuNPs. Two sets of gold nanoparticles (AuNPs) each
is modified with a half ERE segment (v1 and v2) containing 3 bases complementary sticky ends. UV-vis spectra of the complementary mixture of v1-v2 particles
conjugates for (A) AuNPs and (B) AgNPs with increasing amount of ERα in the 50m MKCl-containing protein binding buffer [14] (reproduced with permission from
ref. 12, @ 2010 American Chemical Society).
Qualitatively, the affinity of TFs to different DNA probes could be identified with the degree of change in fluorescence intensity, taken from titration curves, i.e. fluorescent intensity change as a function of TF concentration. The extent of CP quenching merely by dsDNAAuNP defined the baseline for the subsequent calculation of binding constant and stoichiometry. With a “light off” sensor as example with the [protein] dependent fluorescence intensity scales, protein-DNA Kd is calculated through (F0 – F) / (F – Fsat) = (protein]/Kd)n. The Kd was obtained by plotting log [(F0 – F) / (F – Fsat)] versus log [protein], where F0 and Fsatare the relative fluorescence intensities in the absence of protein and in protein saturation respectively. The value of log [protein] at log [(F0 – F) / (F – Fsat)] = 0 equals to the logarithm of the Kd. The slope, n, is the binding stoichiometry of the protein to DNA Figure 4.
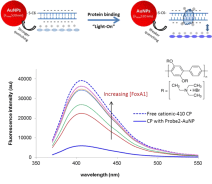
Figure 4: Hybrid sensor using dsDNA-coated AuNPs and a cationic CPE
(Poly[(2,5-bis(2-(N,N-diethylammoniumbromide)ethoxy)-1,4-phenylene)-alt-
1,4-phenylene] for detecting protein (FoxA1) binding to its DNA element.
With this assay again one can determine binding affinity constant and stoichiometry. This assay overcomes the limitation of assay that uses unmodified AuNPs and colorimetric principle. The dsDNAcoated AuNPs is more stable and eliminated nonspecific surface attachment, so that is suitable for studying complex sample and for screening ligand inhibition [18]. The use of fluorimetric principle makes this assay more sensitivity to provide sequence rule with single base resolution [17].
A nanoplasmonic-fluorescent ruler for detection of site specific protein binding to composite dna (Assay IV) [21]
As mentioned earlier in molecular biology and genetics most TFs do not work alone. Often, for gene transcription to occur a number of TFs must bind to DNA regulatory sequences synergistically for example specific protein 1 (Sp1) and estrogen receptor a (ERa) [1]. We construct a gold or silver nanoparticles (AuNPs & AgNPs) supported Fluorescent Intercalator Displacement (FID) assay, termed nanoplasmonic-fluorescent ruler for site specific detection of protein binding to composite DNA of multiple sites (Figure 5A). A 20 nm, 100 nm AuNPs or 80 nm AgNPs is introduced to one end of double stranded DNA (dsDNA). The dsDNA-AuNPs conjugates are saturated with a DNA intercalator, Thiazole Orange (TO). The distance-determined fluorescent quenching and enhancement by mNPs gives each intercalated TO its own contribution to the overall fluorescent intensity according to their distance to the particles. Protein binding at a specific DNA site displaces the respective TO molecules, causing relative fluorescent intensity drop that is of a function of distance. Proteins Sp1 and ERa and their composite DNA, containing two identical Sp1 sites and one ERa site are studied. With the smaller particle (i.e. 20 nm AuNPs) and larger particles (i.e. 100 nm AuNPs and 80 nm AgNPs) as quenching and enhancement profilers, respectively, we can detect preferential binding of Sp1 to one of its sites, identify protein bound, determine the cooperative binding of Sp1 and ERa where conventional FID fails unless a comprehensive set of DNA with respective protein sites being mutated is involved. Figure 5B and 5C are the example of 20 nm AuNPs quenching profiler assisted assay for detecting preferential binding of Sp1 to the Sp1/P site.
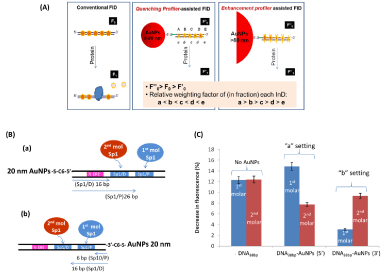
Figure 5: (A) Schematic illustration of conventional FID with even InD emission in dsDNA and metal NPs-coupled FID with uneven InD emission in dsDNA-AuNPs
conjugate. According to their size determined effect the smaller AuNPs in and the larger AuNPs in are termed “quenching profiler” and “enhancement profiler”
respectively. (B) Schematic illustration of two TO/dsDNA-AuNPs20nm settings used to identify preferential binding site of Sp1 at two Sp1/D and Sp1/P sites. The 20
nm AuNPs are attached at the (a)½ ERE site and (b) Sp1/P site, respectively. The bp number refers to the distance of the Sp1 sites to AuNPs (4 bp spacer between
½ ERE and Sp1/D). (C) The relative decrease in fluorescent intensity (at 535 nm) induced by first molar (blue) and second molar of Sp1 (red) binding for dsDNA/
TO without AuNP and fordsDNA-AuNPs/TO composites in “a setting” and “b setting” [21] (Reproduced with permission from ref. 19, @ 2014 WILEY).
AuNPs-based assays for protein-DNA binding developed in other group
DNA cleavage enzyme can digest DNA chains that have also been used in design protein-DNA binding assays. Ou et.al utilized two sets of single stranded DNA (ssDNA)-conjugated DNA to form crosslinked AuNPs network through interparticle DNA hybridization [24]. Protein binding to the preformed dsDNA network protects the DNA linkers from cleavage by exonuclease III (Exo III). As a result, the blue AuNPs aggregates will retains, as a measure of the protein binding. For a simple aggregation assay, in the case when the protein to be studied carries two binding sites to a DNA element, protein binding can be detected by its crosslinking of DNA-conjugated AuNPs, without relying on salt screening and enzymatic DNA cleavage [25]. This concept has been demonstrated for the interaction between the lac repressor (protein) and operator (DNA) and its interplay with the lac operon inducer isopropyl beta- D-1-thiogalactopyranoside (IPTG, which inhibits the interaction between the lac repressor and operator).
Performance Comparison
We have presented four enzyme free AuNPs-based bioassays for studying protein-DNA interactions. Each of them has its own strengths and favorable applications. We regard these assays as a suite of bioassay kits that can collectively provide comprehensive characterization to protein-DNA interactions with different level of complicity, with differential sensitivity resolution. In Table 1 below, we have summarized these assays from the concept, principle, to their ability in providing various characteristics. With the specific objective in main, one can select the right assay to derive the binding characteristics required. In general, the based AuNPs based assay I is fast and easy to operate, but with the limitation for purified protein, and being successful only partially for studying ligand binding effect, for those ligands that do not have affinity to AuNPs. For the assays using DNA-conjugated AuNPs (Assay II and III), since the AuNPs carries a DNA shell, the AuNPs are more stable and exist no nonspecific adsorption from ligand or other proteins. Thus they can be used to study protein from crude samples (like nuclear extract). The assay III is further proved for its feasibility on ligand screening. Due to the fluorimetric detection, which is more sensitive than the colorimetric sensing, assay III can screen sequence selectivity with a single base resolution. Assay IV is a very intelligent design that for the first time DNA intercalator and mNPs are combined for site specific binding measurement. We have proven that most of the assays can provide quantitative measurement of binding affinity.
Assay
I
II
III
IV
Sensing element
Bare AuNPs
DNA-conjugated AuNPs (two sets)
DNA-conjugated AuNPs or AgNPs (one set)
Collaborative sensing element
NIL
Conjugated Polymer
DNA intercalators
Sensing principle
Colorimetric
Fluorimetric
salt induced NPs aggregation
Synergetic effect of salt and DNA base paring induced aggregation
AuNP-CP quenching
Metal NPs-InD quenching
Metal NPs-InD enhancement
Application
Protein-DNA binding
Protein-DNA binding
Protein-DNA binding
Ligand inhibition
Multiple protein-composite DNA
For ligand inhibition study
Yes, but with limitationa
Not demonstrated
Yes
Not demonstrated
Site Specific detection
No
No
No
Yes
Binding Stoichiometry measurement
Yes
No
Yes
Not demonstrated
Table 1: A summary of the four assays from their concept, principle, to their ability in providing various characteristics only possible for the small molecular compounds that do not have affinity to AuNPs and do not intrinsically aggregate the AuNPs [11].
Conclusion
With this review we have demonstrated how powerful of metal nanoparticles and their optical properties in enabling biological analysis, with protein-DNA interaction as an example. Both aggregation induced color change and super quenching can be harnessed as sensing principles, to interrogate protein-DNA interactions with high sequence selectivity resolution and with the capability to determine ligand effect and site specific protein binding. The incorporation of collaborative sensing elements (e.g. conjugated polyelectrolyte and DNA intercalator) can introduce additional versatility in the bioassay design. Moving forwards, we expect that more efforts are put to develop nanoparticles assays for detecting the transcriptional factors alterations and interactions inside the intact cells.
Acknowledgment
Su X would like to thank A*STAR BMRC and JCO fund of 14/1/16/24/008 and 14302FG096 respectively.
References
- Petz LN, Ziegler YS, Schultz J.R, Kim H, Kemper J. K, Nardulli A.M. Differential regulation of the human progesterone receptor gene through an estrogen response element half site and Sp1 sites. Steroid Biochemistry and Molecular Biology. 2004; 88: 113.
- Xiao R, Roman-Sanchez R, Moore DD. DamIP: a novel method to identify DNA binding sites invivo. Nucl Recept Signal. 2010; 16: 8.
- Cheung E, Acevedo ML, Cole PA, Kraus WL. Altered pharmacology and distinct Coactivator Usage for estrogen receptor-dependent transcription through activating protein-1. Proceedings of the National Academy of Sciences. 2005; 102: 559-564.
- Teh HF, Peh WY, Su X, Thomsen JS. Characterization of protein--DNA interactions using surface plasm on resonance with various assay schemes. Biochemistry. 2007; 46: 2127-2135.
- Peh WYX, Reimhult E, Teh HF, Su X, Thomsen JS. Understanding Ligand Binding Effects on Conformation of Estrogen Receptor a -DNA Complex: A Combinational Quartz Crystal Microbalance with Dissipation and Surface Plasmon Resonance Study. Biophysical Journal. 2007; 92: 4415-4427.
- Song HY, Sun S, Prabhakar S, Aung KM, Su X. Study Sequence Rules of estrogen receptor a- DNA interactions using dual polarization interferometry and computational modeling. Anal Biochem. 2013; 433: 121-128.
- Li N, Lu Y, Su X. Nanomaterial-based Biosensors using dual transducing elements for solution phase detection. Particle and Particle System. The Analyst. 2015; 140: 2916-2943.
- Wang J, Qu X. Recent progress in nanosensors for sensitive detection of biomolecules. Nanoscale. 2013; 5: 3589-3600.
- Song SP, Qin Y, Huang Q, Fan CH, Chen H-Y. Functional nanoprobes for ultrasensitive detection of biomolecules. Chemical Society Reviews. 2010; 39: 4234-4243.
- Tan YN, Su X, Liu E, Thomsen JS. Gold-Nanoparticle-Based Assay for Instantaneous Detection of Nuclear Hormone Receptor-Response Elements Interactions. Analytical Chemistry. 2010; 82: 2759-2766.
- Aung KM, New SY, Hong SZ, Tan SK, Cheung E, Su X, et al. Studying fork head box protein A1-DNA interaction and ligand inhibition using gold nanoparticles, electrophoretic mobility shift assay, and fluorescence anisotropy. Anal Biochem. 2014; 448: 95-104.
- Sutarlie L, Aung KMM, Lim MGL, Lukman S, Cheung E, Su X. Studying Protein-DNA Complexes Using Gold Nanoparticles by Exploiting Particle Aggregation, Refractive Index Change, and Fluorescence Quenching and Enhancement Principles. Plasmonics. 2014; 9: 753-763.
- Tan YN, Lai A, Su X. Interrogating Cooperative Interactions of Transcription Factors with Composite DNA Elements using Gold Nanoparticles. Science of Advance Materials. 2014; 6: 1460-1467.
- Tan YN, Su X, Zhu Y, Lee JY. Sensing of transcription factor through controlled-assembly of metal nanoparticles modified with segmented DNA elements. ACS Nano. 2010; 4: 5101-5110.
- Tan YN, Su X, Lee KH. A study of DNA design dependency of segmented DNA-Induced gold nanoparticle aggregation towards versatile bioassay development. RSC Adv. 2013; 3: 21604-21612.
- Seow N, Tan YN, Yung LY, Su X. DNA-Directed Assembly of Nanogold Dimers: A Unique Dynamic Light Scattering Sensing Probe for Transcription Factor Detection. Scientific Report. 2015; 5: 18293.
- Lukman S, Aung KM, Liu J, Liu B, Su X. Hybrid sensor using gold nanoparticles and conjugated polyelectrolytes for studying sequence rule in protein-DNA interactions. ACS Appl Mater Interfaces. 2014; 5: 12725-12734.
- Lukman S, Aung KMM, Lim MGL, Hong S, Tan SK, Cheung E, et al. Hybrid assembly of DNA-Coated gold nanoparticles with water soluble conjugated polymers for studying protein-DNA interaction and ligand inhibition. RSC Advances. 2014; 4: 8883.
- Su X.D, Aung KMM, Lukman S, Liu B. Gold Nanoparticle-Based Forster Resonance Energy Transfer (FRET) Analysis of Estrogen Receptor: DNA Interactions. Estrogen Receptors.
- New SY, Aung KMM, Lim LG, Tan SK, Lu Y, Cheung E, et al. Fast Screening of Ligand-Protein Interactions based on Ligand-Induced Protein Stabilization of Gold Nanoparticles, Analytical Chemistry. 2014; 86: 2361-2370.
- Lukman S, Sutarlie L, Li N, Su X. A Nanoplasmonic-Fluorescent Ruler for Detection of Site-Specific Protein Binding to Composite DNA of Multiple Sites. Particle & particle Systems Characterization. 2014; 31: 1281-1290.
- Su X, Kanjanawarut R. Control of metal nanoparticles aggregation and dispersion by PNA and PNA-DNA complexes, and its application for colorimetric DNA detection. ACS Nano. 2009; 3: 2751-2759.
- Kanjanawarut R, Su X. Colorimetric Detection of DNA Using Unmodified Metallic Nanoparticles and Peptide Nucleic Acid Probes. Analytical Chemistry. 2009; 81: 6122-6129.
- Ou LJ, Jin PY, Chu X, Jiang JH, Yu RQ. Sensitive and visual detection of sequence-specific DNA-binding protein via a gold nanoparticle-based colorimetric biosensor. Anal Chem. 2010; 82: 6015-6024.
- Fang J, Yu L, Gao P, Wei YA, Cai Y. Detection of protein–DNA interaction and regulation using gold nanoparticles. Analytical Biochemistry. 2010; 399: 262-267.