
Research Article
Austin J Biosens & Bioelectron. 2015;1(1): 1005.
Assembly of Gold-Binding Proteins for Biomolecular Recognition
Zareie HM1,2* and Sarikaya M3
1Department of Material Science and Engineering, Izmir Institute of Technology, Turkey
2Microstructural Analysis Unit, School of Physics and Advanced Materials, Australia
3Materials Science and Engineering & GEMSEC, University of Washington, USA
*Corresponding author: Hadi M. Zareie, Department of Material Science and Engineering, Izmir Institute of Technology, Urla, Izmir, 35430 Turkey.
Received: October 23, 2014; Accepted: February 04, 2015; Published: February 09, 2015
Abstract
Controlled binding and assembly of proteins onto inorganics is at the core of biological materials science and engineering with wide ranging applications. Here we demonstrate ordered assembly of genetically-engineered inorganicbinding polypeptides on an atomically-flat solid surface. We used a 3-repeat, 14 amino acid Gold-Binding Protein (GBP1) that forms a monolayer-thick film with nano structured domains on the gold surface. The protein film conforms into 3-fold symmetry, commensurate with Au (111) lattice, suggesting crystallographic recognition. The GBP1 was selected for its specific affinity to gold by using cell-surface display. The engineered proteins could have significant potential impact by providing self-assembled functional molecular substrates in nanoand bio-technologies. We demonstrate directed assembly of ferritin (target) onto streptavidin (probe) conjugated to biotinylatedGBP1, and quantitatively analyze the results.
Keywords: Gold-Binding Protein (GBP); Self-assembly; Atomic Force Microscopy (AFM)
Abbreviations
GBP1: Gold-Binding Protein; SAMs: Self-Assembled Monolayers; MD: Molecular Dynamics; SPR: Surface Plasmon Resonance; bio- GBP1: biotinylated GBP1; SA: Streptavidin; GEPI: Genetically Engineered Polypeptides for Inorganics
Introduction
The attachment of biomolecules, in particular proteins, onto solid supports is fundamental in the development of advanced biosensors, bioreactors, and many diagnostics such as those used in cancer therapeutics [1-5]. The realization of biology-inspired materials technologies depends on understanding the nature of the chemical and physical interactions between macromolecules and biominerals or human-made inorganic materials [3]. Macromolecular interactions at solid surfaces play key roles in implants and hard-tissue engineering [4], and proteins adsorbed onto substrates are used to build protein microarrays suitable for modern proteomics [5]. With a new approach, we show self-assembly of a genetically engineered Gold-Binding Protein (GBP1) as a monomolecular film forming self-patterned domains, immobilize a probe protein onto it, and show its utility in target recognition. Our results demonstrate unique advantages offered by molecular biomimetics [6] including surface recognition, self-assembly, and genetic engineering, in creating functional molecular substrates for potential nanobiotechnological applications.
Combinatorial genetic techniques (both cell-surface display and phage display) permit isolation of specific recognition elements for surfaces, including those not recognized by natural proteins, in the absence of a priori prediction of necessary structures [7-8]. Recently, a number of inorganic binding polypeptide sequences have been selected using display technologies [9-13]. In our experiments, GBP1, a gold-binding protein, was used with an amino acid sequence MHGKTQATSGTIQS, repeated three times to increase its affinity [9,11]. Selection of gold binders was designed to produce modular, independently folding metal recognition sequences. Gold-binding sequences had been isolated as extracellular loops of maltoporin [9]. Many proteins bind tightly to gold at low salt concentrations; however the engineered proteins were selected to bind at higher salt concentrations [9,11]. To improve the binding activity, multiple repeats (up to 11) of this sequence was generated. The binding motif does not contain cysteine which is known to form a covalent thiol linkage with gold, the linkage to the gold surface in Self-Assembled Monolayers (SAMs) [14]. In our experiments, strong binding activity required at least 3 repeats and most of our current experiments were carried out using this protein.
Material and Methods
GBP1 self-assembly
The gold sample was prepared following the well-known protocol frequently used in the preparation of Self-Assembled Monolayer (SAM). Atomically-flat 0.5 to 1.0 μm diameter gold grains were produced, textured about Au [111] axis (Figure 1A and B). For the assembly of the proteins, the gold substrates were immersed into 1 mg/ml GBP-1 solution for a period from a few minutes to few hours and days. The samples were rinsed with the buffer solution first and then with de-ionized water to remove physisorbed multilayers to ensure that the remaining molecules were strongly bound to the surface (see SPR results below). After a sufficient period of incubation, the gold surfaces were examined using an AFM in air.
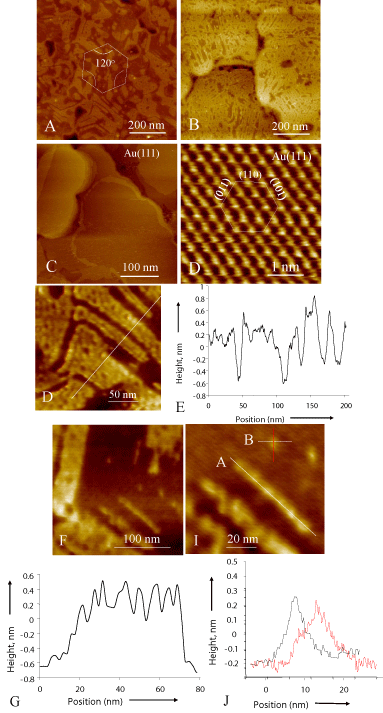
Figure 1: Assembly of GBP1 characterized by SPM. (A) and (B) are AFM
images showing ordered GBP1 domains on Au (111). The STM images of the
gold surface reveal flat grains (A) textured about <111> in 6-fold symmetry
(B). The edges of the self-assembled protein domain in (A and B) have either
60o or 120o angle, suggesting possible alignment along crystallographic
directions on Au (111). (E - J) Analyses of detailed structure of GBP1
domains. The image (E) displays domains in partially assembled proteins on
Au(111) surface and (F) is the corresponding line profile taken across several
domains with straight boundaries revealing a monolayer thickness across
all the domains.The AFM image in (G) reveals a narrow and straight single
molecule-thick domain, probably aligned along a 6-fold crystallographic axis,
and (H) is line profile along it, displaying peaks that are likely to correspond to
individual proteins. (I) is an image of a single protein and (J) is its line profiles
taken orthogonally across the molecules giving approximate dimensions.
Scanning probe microscopy
SPM was performed using a Nanoscope-III system (Veeco Instruments, Santa Barbara, CA) operated in air at room temperature. Freshly-cut Pt-Ir tips (0.2 mm diameter) were used and STM images were obtained with typical tip-sample bias voltages of 50 mV and tunneling currents 1nA. AFM imaging was conducted in tapping mode with 512x512 data acquisition at a scan rate of 1.4 Hz using oxide-sharpened Si3N4 tips with integrated cantilevers of the nominal spring constant of 0.38 N/m. Image analyses were carried out using SPIP software (Image Metrology ApS, Lyngby, Denmark).
Surface plasmon resonance spectroscopy
The SPR instrument used was based on a planar prism (Kretchmann) configuration, described and characterized elsewhere. It can detect changes in bulk solution refractive index down to ~2x10-6 (corresponding to ~1x10-3 protein monolayers for a system). Whitelight is directed at the gold-coated substrate through the prism at a fixed angle (78° from normal), and adsorption-induced changes in the Refractive Index (RI) near the gold sensor surface are observed by automated monitoring of the changes in wavelength where the SPR reflectance minimum occurs. These shifts are converted into coverage's of the proteins using a formalism for quantitative SPR which involves a sensitivity calibration based on the sensor response to the changes in the bulk RI, an exponential probe-depth estimated from Frensel equations, and the known RI for these class of proteins.
Probe/target molecules, and biotinylation
Strepavidin (Calbiochem) was dissolved in water (1mg/mL). The water used was purified by reverse osmosis followed by a Millipore unit (18 mW resistivity). Both GBP1 and ferritin (horse spleen, from Sigma) were biotinylated through the lysine (K) amino groups using sulfusoccinimidyl-6 (bioyinamido) hexonate (Sulfo-NHS-LC-biotin, from Pierce). The biotinylation reactions were performed in 100 mM phosphate buffer (pH ~ 7.5) at room temperature for 2 hours. The mol ratios of biotin to GBP1 and ferritin were 3 and 5, respectively. After the reaction, biotinylated ferritin was purified using a size exclusion column (PD-10, Amersham) equilibrated with 10mM phosphate buffer.
Result and Discussion
We demonstrate the dramatic effect of assembly of GPB1 on Au (111) in Figure 1. Gold substrates containing large and atomicallyflat (111) oriented grains (Figure 1A and B) were prepared following the well-known protocol used in the preparation of Self-Assembled Monolayer (SAM) [14,15]. Even after 20-minute incubation in protein solution, monomers form well-defined domains with fairly straight edges and sharp corners that progressively spread out to cover the gold surface (Figure 1 C and D). These nanoscale domain architectures could be indicative of the GBP1 conforming with the crystallography of the Au (111) surface. Measurements of the angles among domain edges produce 60o or 120o-separation. The ordered assembly of the proteins might suggest that GBP1 molecularly recognizes the Au (111) surface. A recent study, using Molecular Dynamics (MD), indicates that GBP1, with an antiparallel β-sheet structure (inset in 1A), recognizes gold surface via OH- binding [16]. It is likely that the hydroxyl, together with amine, ligands on GBP1 recognize the atomic lattice of gold, aligning the molecule along the variants of a six-fold axis on the (111) surface (analysis not shown). The AFM images are analyzed further to establish molecular details of the ordered structures of the GBP1. A line profile (Figure 1E) over several parallel domains (Figure 1F) shows a 0.6±0.2 nm height difference with the substrate surface, corresponding to the monolayer thickness of the adsorbed polypeptide. Also, a profile taken along a narrow domain corresponds to molecular units (Figure 1G and H) that appear to have lined up forming a linear domain. Similarly, measurements of a single molecular unit (captured in Figure 1I and J) produce an approximate size of (3±2 x 7±2 nm). These dimensions are in agreement with those predicted by the MD studies for the 3-repeat GBP1 [16].
Dynamics of the surface coverage and binding affinity of GBP1 onto gold surface (Figure 2A) were studied using Surface Plasmon Resonance (SPR) spectroscopy. The shift in SPR wavelength is related to protein adsorption onto gold surface with respect to a baseline shift [17,18]. In our experiments, a sharp increase in SPR shift represents a rapid assembly of GBP1 onto sensor surface. The decline of the SPR profile is due to the removal of unbound or physisorbed proteins upon rinsing. Indicative of stable coverage of the surface, the final buffer rinse drops the wavelength shift to a specific value (4.529 nm) corresponding to 1.49x1012 molecules/cm2. This value compares well with the calculated one, 5x1012 molecules/cm2, and assuming full coverage and based on the MD-predicted shape and dimension of the GBP1 molecule. Time-sequence imaging of the adsorption process was also carried out using AFM to monitor structural changes during the protein assembly process on the substrate surface (Figure 2 B - D). Although early on (within 5-10 minutes) proteins aggregate forming random and branched clustering (Figure 2B), after about 15 minutes of incubation, ordered assembly of protein domains form with straight boundaries and sharp corners (Figure 2C). A possible mechanism for the ordering could be that, following the adsorption onto solid surface; the proteins could reorganize through a combination of forces involving Au-lattice recognition and intermolecular interactions among protein units. These studies reflect that nearly full ordered coverage of the GBP1 obtained within about an hour (Figure 2D).
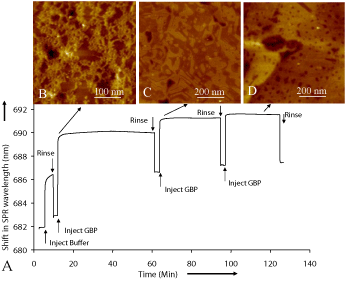
Figure 2: SPR study of GBP1 assembly on gold. (A) The profile is a shift in
the SPR wavelength versus the time of immersion of the gold substrate in
the protein solution. A rapid increase in the profile at the beginning reflects
that the protein assembly process starts rapidly, and saturates quickly. The
sample was periodically taken out of the solution and flushed thoroughly
with the buffer solution followed by cleaning with DI water to wash away
physisorbedmolecules; this procedure corresponds to the drops in the SPR
profile. The final value of the shift corresponds to the fully covered surface
by the proteins giving a value for saturation of adsorption. (B-D) The AFM
images of the samples (proteins on Au(111)) correspond to the immersion
times of the SPR data (taken using the polycrystalline gold), suggesting that
assembly process starts with a rapid but random aggregation of the proteins
with a branched structure, which then reorganizes into ordered monolayerthick
two-dimensional domains.
We next demonstrate the utility of the self-assembled GBP1 as a functional molecular erector set by using GBP1 and immobilize a probe protein onto it. The assembly sequence is schematically shown in Figure 3A which is experimentally demonstrated using SPR (Figure 3B) and confirmed with AFM analysis (Figure 3C-E). First, biotinylated GBP1 (bio-GBP1) is assembled onto gold surface, with the same ordered assembly as GBP1 alone, possibly indicating that biotin-terminated surface is exposed (Figure 3C). Streptavidin (SA, the probe molecule [19]) was then immobilized onto this ensemble with a surface coverage of ~ 2.1x1012 molecules/cm2 (Figure 3D). SA does not bind to native GBP1 (non-biotinylated) in a controlled experiment (not shown). We next immobilized biotinylated ferritin (FE, a model target protein [20] onto this functional molecular substrate to assess that the immobilized SA has retained its specificity and biological activity (Figure 3E). The value of the SPR wavelength shift (7.0 nm) corresponds to a surface coverage of 3.14x1011 molecules/cm2, i.e., nearly 95% of the SA binding sites is occupied. This is a highly efficient immobilization and compares well with the self-assembled biotin-terminated alkylthiols on gold surface that are traditionally used for biosensor applications immobilizing probe proteins (e.g., SA) for target protein [21] or DNA selection [22].
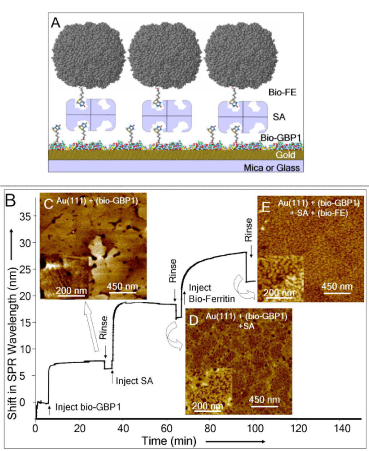
Figure 3: Demonstration of GEPI-based protein film as a molecular erector
set. (A) A schematic of the assembled molecules on gold: bio-GBP1:
biotinylated 3-reapeat gold-binding protein; SA: streptavidin, and Bio-FE:
biotinylated ferritin. (B) The analyses of multiple assembly/immobilization
processes studied by SPR and corresponding immobilized structures imaged
via AFM (C-E). Surface saturation in each case is evidenced by no shift in
SPR wavelength, i.e., flat portion of the profile. The assembly processes
involve a sequence of three stages: (I) Bio-GBP1 is assembled onto Au(111)
forming the same type of domains as in the pure GBP1 case giving a threefold
symmetry (C); (II) SA (the probe protein) is immobilized onto bio-GBP1
(D); and (III) Bio-FE (the target protein) is immobilized onto SA+Bio-GBP1
(E).
Our protein, GBP1, represents a new class of biological molecules [9-13], genetically engineered polypeptides for inorganics (GEPI) that are combinatorially selected to bind to specific inorganics. The ordered assembly of a GEPI on inorganic surfaces could have a significant impact in molecular biotechnology applications offering several novel practical advantages. Firstly, the number density of molecules could be controlled for a given surface; an essential parameter to accurately calculate probe density, which, in traditional self-assembled systems, has only been possible to estimate [5,18]. By using a GEPI, one could utilize multiple sites for fusion with a desired ligand, several ligands of the same type, or several different ligands, increasing binding efficiency and versatility; this is unlike an alkylthiolate molecule, which has just one single available active site for fusion of a probe molecule. The kinetics of the GEPI assembly is fast (minutes) and robust, stability lasting months (data not shown), both essential parameters for practical applications. The GEPI-based molecular erectors are biology-friendly (assembled at room temperature in aqueous solutions at pH 7) providing applicability in a wide-range of in vivo cases. This is contrary, e.g., to alkylthiolates that are usually assembled in non-biological solutions and, therefore, could be a setback in some applications [5,22]. The fusion of the probe could not only be accomplished chemically in thiol- and silane-based systems [21,22]; genetic fusion is also possible, and advantageous, in the GEPIbased systems. The possibility to attach a given biomacromolecule (enzyme or antigen) directly onto a GEPI via genetic fusion give them an uncontested advantage as these approaches reduce possible chemical-binding mistakes, increase purity, and, most importantly, allow in vivo genetic manipulation [5,21,22]. Furthermore, GEPIbased molecular substrates could also be used for selected assembly and controlled organization of nanostructured units, e.g., inorganic nanoparticles (data not shown) and nanowires on surfaces providing a means for molecular fabrication of functional inorganics, a utility in nanotechnology [1].
Acknowledgement
Technical help by D. Heidel and V. Bulmus, and the use of SPR facilities of S. S. Yee (all at University of Washington) are acknowledged. The model of GBP1 (Figure 1A) was based on MD studies, provided by R. Braun and K. S. Schulten, Beckman Institute, University of Illinois, Urbana-Champaign. This work was supported by US-ARO through a DURINT program.
References
- Niemeyer CM. Nanoparticles, Proteins, and Nucleic Acids: Biotechnology Meets Materials Science. Angew. Chem. Intl. Edit. 2001; 40: 4128-4158.
- Talbot J, Tarjus G, Van Tassel PR, Viot P. From car parking to protein adsorption: an overview of sequential adsorption processes. Coll. & Surf. A. 2000; 165: 287-324.
- Weiner S, Addadi L. Design strategies in mineralized biological materials. J. Mater. Chem. 1997; 7: 689-702.
- Sakiyama-Elbert SE, Hubbell JA. Functional biomaterials: Design of novel biomaterials. Ann. Rev. Mater. Res. 2001; 31: 183-201.
- Cutler P. Protein arrays: the current state-of-the-art. Proteomics. 2003; 3: 3-18.
- Ball P. Life's lessons in design. Nature. 2001; 409: 413-416.
- Hoess RH. Protein design and phage display. Chem Rev. 2001; 101: 3205-3218.
- Wittrup KD. Protein engineering by cell-surface display. Curr Opin Biotechnol. 2001; 12: 395-399.
- Brown S. Metal-recognition by repeating polypeptides. Nat Biotechnol. 1997; 15: 269-272.
- Kjaergaard K, Sørensen JK, Schembri MA, Klemm P. Sequestration of zinc oxide by fimbrial designer chelators. Appl Environ Microbiol. 2000; 66: 10-14.
- Brown S, Sarikaya M, Johnson E. A genetic analysis of crystal growth. J Mol Biol. 2000; 299: 725-735.
- Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000; 405: 665-668.
- Naik RR, Brott LL, Clarson SJ, Stone MO. Silica-precipitating peptides isolated from a combinatorial phage display peptide library. J Nanosci Nanotechnol. 2002; 2: 95-100.
- Whitesides GM, Mathias JP, Seto CT. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science. 1991; 254: 1312-1319.
- Schreiber R. Structure and growth of self-assembled monolayers. Prog. Surf. Sci. 2000; 65: 151-156.
- Braun R, Sarikaya M, Schulten K. Genetically engineered gold-binding polypeptides: structure prediction and molecular dynamics. J Biomater Sci Polym Ed. 2002; 13: 747-757.
- Jung LS, Campbell CT, Chinowski TM, Mar MN, Yee SS. Quantitative interpretation of the resonance of surface plasmon resonance sensors to adsorbed films.Langmuir. 1998; 14: 5636-5648.
- Perez-Luna HV, O'Brien MJ, Opperman KA, Hampton PD, Stayton PS, Klumb L, et al. Molecular recognition between genetically engineering streptavidin and surface-bound biotin. J. Amer. Chem. Soc. 1999; 121: 6469-6478.
- Jung LS, Nelson KE, Stayton PS, Campbell CT. Binding and dissociation kinetics of wild-type and mutant streptavidins on mixed biotin-containing alkylthiolate monolayers.Langmuir. 2000; 16: 9421-9432.
- Hodneland CD, Lee YS, Min DH, Mrksich M. Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands. Proc Natl Acad Sci U S A. 2002; 99: 5048-5052.
- Panda S, Sato TK, Hampton GM, Hogenesch JB. An array of insights: application of DNA chip technology in the study of cell biology. Trends Cell Biol. 2003; 13: 151-156.
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996; 1275: 161-203.