Research Article
Austin J Biotechnol Bioeng. 2014;1(1): 6.
High Prevalence of Antibiotic Resistance in Escherichia coli Isolated from Fecal Sample of Cows and Assessment of Antibacterial Efficacy of Indigenous Medicinal Plants from Assam, India
Borah VV, Bora P, Roy M and Saikia KK*
Department of Bioengineering and Technology, Gauhati University, India
*Corresponding author: Saikia KK, Department of Bioengineering & Technology, Gauhati University, Gopinath Bordoloi Nagar, Guwahati - 781014, Assam, India.
Received: June 27, 2014; Accepted: July 21, 2014; Published: July 23, 2014
Abstract
Cow dung is the most common organic manure in agriculture in Assam, India. Escherichia coli were isolated from cow dung samples in between January to May, 2013. The isolates were tested for antibiotic sensitivity to multiple antibiotics and production of ESBLs as per the NCCLS guidelines, 2002 and genotypically through PCR for the presence of bla genes. E. coli were isolated in 80% of the total 100 samples, of which 15% showed resistance to multiple drugs. Of these 80 isolates, 59 (73.75%) were found to be resistant to at least one of the 3GCs. PCR data revealed blaTEM in 25/28 isolates, blaSHV in 6/28 isolates, blaCTX-M in 8/28 isolates and blaAmpC in 27/28 isolates; both blaTEM and blaSHV were present in 21.42%, blaTEM and blaCTX-M in 28.57%, blaSHV and blaCTX-M in 10.71%. ESBLs are the main cause of resistance to beta-lactam antibiotics in E. coli. ESBL mediating resistance to 3GC was found in 35% of isolates. The leaves of Holarhenna antidysenterica wall, and flowers of Woodfordia fructicosa (L.) Kurz were collected at the same time and its antibacterial activity was assessed against antibiotic resistant isolates. Methanolic crude extracts from these medicinal plants exhibited inhibitory activity against TEM+SHV encoding enteric isolates from cow. Detection of enteric ESBL producers in cows in Assam, India in high numbers imply the risk of dissemination of antibacterial resistance into the environment being relatively high and thus, warrant preventive measures.
Keywords: Cow dung; SHV; TEM; CTX-M; Medicinal plants
Abbreviations
ESBL: Extended Spectrum β-lactamases; IMViC: Indole, Methyl red, Voges Proskauer and Citrate utilization test; dNTP: deoxynucleotides; TEM: Temoniera; SHV: Sulphydryl variable; CTX-M: Cefotaximase, Munich; DMSO: Dimethyl Sulphoxide; 3GC: 3rd Generation Cephalosporin; NCBI: National Centre for Biological Information; BLAST: Basic Local Alignment Search Tool; NCCLS: The National Committee for Clinical Laboratory Standards
Introduction
Antimicrobial resistance has been recognized as one of the world's most pressing public health problems [1,2]. The intensive use and, particularly, the misuse of antibiotics have led to the development and selection of resistant bacteria in different settings. Beyond the use of drugs for therapeutic purposes in the human and animal settings, antibiotics are also used extensively as prophylactic agents and as animal growth promoters in agriculture, except in European Union countries, where their use is banned since 2006 in animal feed [3]. Therefore, resistant bacteria are not only confined to the human clinical setting, such as hospitals, where they were first recognized and studied. They have also increased significantly in the community and in both farm and companion animals. Animals may act as reservoirs of resistant bacteria that can be transmitted to humans, or vice versa, by direct contact or indirectly, via the food chain [3-6]. Moreover, the growth of global trade and travel allows resistant microorganisms to be spread rapidly to distant countries and continents [7]. The consequences of antimicrobial resistance represent a growing threat. When infections often fail to respond to standard treatments, they result in prolonged illness and greater risk of death, and more chances for the resistant microorganism to spread. Furthermore, the costs associated with the length of hospitalization and the use of last generation antibiotics are significantly increased [8,9]. The use in human medicine of third generation cephalosporin's (3GCs, e.g. cefotaxime, ceftazidime, ceftriaxone) is generally believed to have been a major selective force in the emergence of extendedspectrum β-lactamases (ESBLs) [8]. Shortly after the introduction of cefotaxime, transferable plasmid-mediated resistance was noted, initially in Germany, with the identification of a mutant β-lactamase SHV-2 that conferred high-level resistance to all of these agents [8]. The following year, in France, a similar mutated variant of TEM β-lactamase (TEM-3) was observed [8] and this led to the adoption of the term Extended Spectrum β-lactamase (ESBL). The small but gradually increasing use of 3GCs in food animal production may be linked to the recent emergence of ESBLs in bacteria associated with cattle, poultry and pigs [10]. It is speculated that emergence of ESBL bacteria in food producing animals may present a risk of resistant strains being transmitted to humans through the food chain. Hence, it asserts importance of studying the prevalence of ESBLs in the veterinary population and their dissemination through the food chain to humans, if any [10].
In the North-Eastern part of India, particularly Assam, cow is the primary source of milk. Hence it is important to identify the most commonly infecting pathogens as well as commensal microorganisms and their resistance patterns to the commonly prescribed antibiotics, as well as molecular mechanism of resistance in resistant isolates. Production of ESBLs is the most common mechanism of resistance to 3G Cephalosporin in Gram negative bacteria. This study was designed with the aim of analyzing the antibiotic susceptibility pattern of E. coli isolated from cow-dung in Guwahati area of India. ESBL producers were detected both phenotypically as well as genotypically. The findings will aid in surveillance study and also act as a guide for prophylactic therapy.
In vitro antimicrobial activity of locally available medicinal plants Holarhenna antidysenterica Wall and Woodfordia fructicosa (L.) Kurz were selected based upon traditional knowledge gathered by interviewing local practitioners were assessed against the ESBL producing enteric isolates. While Woodfordia fructicosa (L.) Kurz finds its application in treating ailments that may be of bacterial origin, for example boils, diarrhea, dysentery, fever, cough, menstrual disorders, urinary disorders, wounds, swellings, cuts, skin diseases [11]. Bark of Holarrhena antidsenterica Wall is used in Ayurveda as an anti-microbial, anti-inflammatory and analgesics, other useful parts used as medicine are root and leaf. In addition the plant has been reported to possess antihelminthic, appetizing, antidiarrhoel and astringent properties [12]. This plant is found throughout tropical India and is an important plant used in indigenous systems of medicine as a remedy for bronchitis, hematuria, spermatorrhoea, epilepsy, asthma, piles, leprosy, eczema, diarrheal, fevers and jaundice [13]. Medicinal plants can be a source of chemical compounds which are able to restrict or stop the growth of microorganisms thereby finding immense application in pharmaceutical research. There is an ever-increasing demand for plant-based therapeutics in both developing and developed countries. Compounds of plant origin are believed to be safe without side effects [14].
Materials and Methods
Ethical clearance
Ethical clearance was obtained from Institutional Ethical Committee of Gauhati University.
Collection of bacterial samples
One hundred cow-dung samples were collected from different cowsheds from various locales within Guwahati region for a period of four months (January to May 2013). The samples were collected into sterile zip lock bags and transferred to the laboratory immediately. The isolation of bacteria was carried out within 4-10 hours from the time of collection.
Isolation and identification
One mg of the sample was added to 10ml of autoclaved saline distilled water and vortexed. An aliquot of 1.0 mL was transferred into the next test tube containing 9.0mL of the sterile distilled water and diluted serially in one-tenth stepwise to 10-2 dilution. With a sterile loop this suspension was streaked on a MacConkey agar and incubated at 37°C for 24 hours. Pink, dry, irregular shaped colonies inferred as E. coli were selected for the study. Further, the E. coli isolates were confirmed by Gram's staining and by standard biochemical test (IMViC tests) [15].
Antibiotic susceptibility profile
The antibiotic susceptibility was determined by the Kirby Bauer's disc diffusion method on Mueller-Hinton agar by inoculating plates with a bacterial suspension compared with a 0.5 McFarland standard, according to the NCCLS guidelines for veterinary samples, 2002 [16]. The antibiotics tested were - Erythromycin (15 μg), Imipenem (10 μg), Kanamycin (30 μg), Co-trimoxazole (25 μg), Ampicillin (10μg), Chloramphenicol (30 μg), Aztreonam (30 μg), Cefotaxime (30μg) and Ceftazidime (30μg) (HiMedia Laboratories Pvt. Ltd., Mumbai, India).
Phenotypic detection of ESBL Producers
All isolates were tested for the detection of ESBL production by the confirmatory method of NCCLS guidelines for veterinary samples, 2002 [16] using cefotaxime (30 μg) and ceftazidime (30 μg) and a disc of cefotaxime plus clavulanic acid (30/10 μg) and ceftazidime plus clavulanic acid (30/10 μg) (HiMedia Laboratories Pvt. Ltd., Mumbai, India) placed at a distance of 20 mm on a lawn culture (0.5 McFarland inoculum size) of suspected ESBL producing clinical isolate on Mueller-Hinton Agar (Hi-Media, Mumbai). Escherichia coli ATCC 25922 were used as the negative control and Klebsiella pneumoniae ATCC 700603 was used as the ESBL positive control. ESBL production was inferred if the inhibition zone was ≥5 mm towards the cefotaxime plus clavulanic acid disc or ceftazidime plus clavulanic acid disc in comparison to the third generation cephalosporin disc alone. Only those isolates identified phenotypically as ESBL producer were selected for genotypic detection of ESBL encoding genes.
Genotypic detection of ESBL encoding genes
To 100μL of sterile Millipore water 4 to 5 colonies of the overnight bacterial culture grown on Luria Bertani Agar (HiMedia Laboratories Pvt. Ltd., Mumbai, India) was taken with a loop. This suspension was boiled at 100°C in a water bath for 10 minutes. Equal volume of Chloroform: isoamyl alcohol (24:1 v/v) was added and the samples were then centrifuged at 12000 rpm for 10 minutes. The upper layer was used as the crude DNA for PCR amplification [17].
PCR analysis was carried out for detection of β-lactamase encoding genes - TEM, SHV, CTX-M and AmpC. Primers obtained were - for blaTEM 5'-CTTCCTGTTTTTGCTCACCCA-3' and 5'-TACGATACGGGAGGGCTTAC-3', for blaSHV 5'-TCAGCGAAAAACACCTTG-3' and 5'-TCCCGCAGATAAATCACC-3' [17], for blaCTX-M 5'-ATGTGCAGYACCAGTAARGT-3' and 5'-TGGGTRAARTARGTSACCAGA-3' [18], and for blaAmpC5'- AATGGGTTTTCTACGGTCTG-3' and 5'-GGGCAGCAAATGTGGAGCAA-3' [19].
Amplification reactions were performed in 25uL volume containing 250 pg of template DNA. For TEM, SHV, CTX-M and AmpC amplification the reaction volume contained 1X PCR buffer, 0.8 mM of MgCl2, 0.2 mM dNTP mix (Invitrogen, North America), 1 μl of each primer (10 μM) with 1 U Taq DNA Polymerase (Invitrogen, North America) per tube. K. pneumoniae ATCC 700603 was used as the positive control for SHV and for TEM; CTX-M and AmpC inhouse positive controls confirmed by PCR-sequencing were used. All PCR amplifications were carried out in Veriti Thermal Cycler (Applied Biosystems, USA).
The cycling conditions for amplification were as follows: for SHV gene, initial denaturation at 94°C for 2 min and 30 cycles of 30 sec at 94OC, 30 sec at 46°C and 45 sec at 72°C, followed by 5 min at 72°C. For TEM gene initial denaturation of 2 min at 94°C and 30 cycles of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C, followed by 7 min at 72°C. For CTX-M gene, initial denaturation at 94°C for 3 min and 30 cycles of 45 sec at 94°C, 30 sec at 48°C and 60 sec at 72°C, followed by 5 min at 72°C and for AmpC gene initial denaturation at 95°C and 30 cycles of 30 sec at 95°C, 30 sec at 51°C and 1 min at 72°C, followed by 2 min at 72°C.
The resulting PCR products were analyzed by electrophoresis with Ethidium Bromide supplemented 1.5 per cent agarose gel in Tris-Acetate-EDTA buffer. The electrophoresis was run at 80-100 Volt till the sample reached 3/4th of the gel. The gels were observed and photographed in a Gel Documentation system (Gel logic, Carestream). A molecular weight standard (Quick-load™100bp DNA ladder; Invitrogen, North America) was included on each gel.
Two PCR amplicon samples for each gene were sequenced and compared to sequences available in National Centre for Biological Information (NCBI) through BLAST program.
Screening of plant antimicrobials
Flowers of Woodfordia fructicosa (L.) Kurz, and leaves of Holarhenna antidysenterica Wall. Were collected from Guwahati region, washed, shade dried and finely grounded to powder. The plant powder was used for the preparation of crude methanolic extract by soxhlet extraction [20] and aqueous extract was prepared by maceration in autoclaved distilled water [21]. The extracts were allowed to dry and then stored at 4°C until further use. Dimethyl Sulfoxide (DMSO) was used to dissolve the extracts to test the antibacterial efficacy which was done by well diffusion method on Muller Hinton Agar [22]. The inoculated plates were incubated for 16-18 hours at 37°C and the zone of inhibition was recorded. The presence of any measure of a zone of clearance indicated antibacterial activity of the plant extract.
Results and Discussion
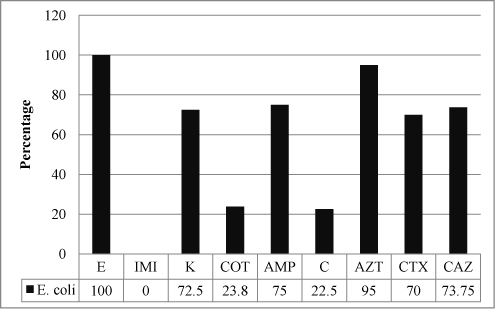
A total of 80 non-duplicate bacterial isolates were cultured on MacConkey Agar from the 100 fecal samples collected and were identified to be E. coli. All the 80 isolates, in our study, were tested for susceptibility to antibiotics and β lactam-β lactamase inhibitor combinations as per NCCLS guidelines, 2002. These isolates tested showed resistance to Erythromycin - 80/80 (100%), Aztreonam - 76/80 (95%), Ampicillin - 60/80 (75%), Ceftazidime - 59/80 (73.75%), Kanamycin - 58/80 (72.5%) and Cefotaxime - 56/80 (70%) (Figure 1).
Figure 1: Bar Diagram showing the percentage resistance of all the enteric E. coli isolates to the antibiotics tested (Key - E - Erythromycin, IMP - Imipenem, K - Kanamycin, COT - Co-trimoxazole, AMP - Ampicillin, C- Chloramphenicol, AT - Aztreonam, CTX - Cefotaxime, CAZ - Ceftazidime).
While the ESBL producing isolates tested showed a high degree of resistance to Erythromycin-28/28 (100%), Aztreonam-25/28 (89.28%), Ampicillin - 25/28 (89.28%), Kanamycin-21/28 (75%), Cefotaxime-24/28 (85.71%) and Ceftazidime-28/28 (100%). The susceptibility profile of Imipenem 80/80 (100%), Co-trimoxazole 61/80 (72.5%) and Chloramphenicol 62/80 (77.5%) indicated to be the most effective antibiotics. Of the 80 isolates, 59 (73.75%) were found to be resistant to at least one of the 3GCs.Of these 59 isolates thus tested for ESBL production, 28 were inhibited by ceftazidime/clavulanic acid. These isolates were tested by placing the ceftazidime and ceftazidime/clavulanic acid discs at a distance of 20 mm and a > 5 mm increase in the zone diameter towards cefotaxime/clavulanic acid was observed as per NCCLS guidelines, 2002, of these 28 isolated, 8 were also inhibited by cefotaxime/clavulanic acid. Thus, among these 59 isolates, 28 (47.45%) were found to be ESBL producer by Doubledisc synergy test. Hence, out of the 80 isolates screened for ESBL production, 28 (35%) were found to be ESBL producers.
To understand the antibiotic resistance changes over a period of time, a study carried out by the Centre for Disease Control showed a significant increase in resistance in E. coli isolates from animals to 11 of 15 antimicrobials inclusive of Ampicillin, Tetracycline and Streptomycin [22]. Prevalence of ESBL was more in females (85.71%, 24 out of 28) than in males (14.29%, 4 out of 28). Of the 80 isolates, 59 (73.75%) were found to be resistant to at least one of the 3GCs. Overall high resistance was observed against commonly prescribed antibiotics. ESBL mediating resistance to 3GC was found in 35% of isolates. The prevalence rate is higher than the reported figure of E. coli in France (5.8%, 2008) [23] and Turkey (1.25%, 2008) [24]. This prevalence appeared to be higher than several studies; for example Duan and others, 2006 [25] reported a 3.1% prevalence ofESBL producers among E. coli isolates from cattle in Hong Kong; it was found that1.5% resistance to cefotaxime in Enterobacteriaceae members isolated from cattle in Japan [26]. Thus it can be concluded that the rate of occurrence of ESBLs has vastly increased in the last few years.
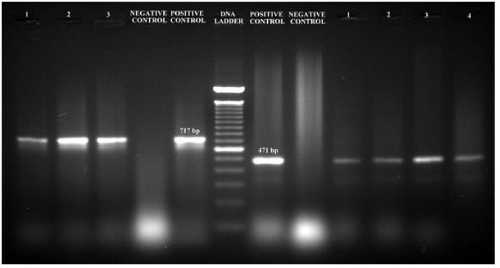
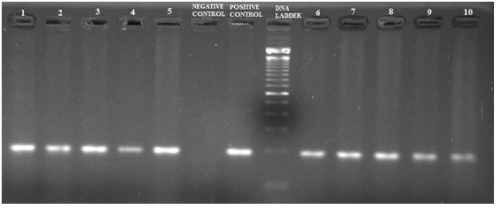
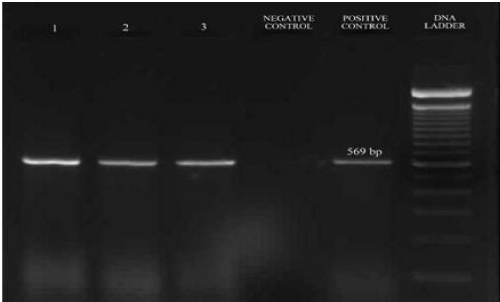
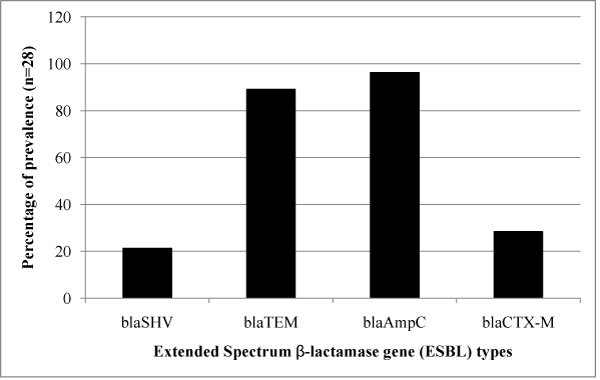
The ESBL positive strains confirmed phenotypically were further tested for the presence of blaSHV, blaTEM, blaCTX-M and blaAmpC genes to identify their resistance mechanism. The amplified product of blaSHV and blaTEM gene had a fragment size 471bp and 717bp corresponded to 100bp molecular weight marker and the positive control (Figure 2). Of the 28 samples, 6 (21.42%) were positive for blaSHV, 25 (89.29%) were positive for blaTEM; presence of both blaTEM and blaSHV genes in 6 (21.42%) isolates and 27 (96.42%) were positive for blaAmpC with a fragment size of 192bp (Figure 3) while 8 (28.57%) were positive for blaCTX-M with a fragment size of 569 bp (Figure 4); a comparison of the presence of two or more ESBL genes in a single isolate is given as Table 1. Figure 5 shows the bar diagram representation of ESBL genes present in the enteric E. coli isolates from cows. It is important to detect the underlying genetic behavior for a phenotypically confirmed ESBL producing isolates to understand the present scenario of dissemination of CTX-M, TEM, SHV or AmpC genes in a veterinary population and whether the trend changes overtime needs to be monitored. High rates of ESBL producers carrying CTX-M, TEM and SHV genes in food animals and the high genetic diversity among these isolates have been reported [27] while the presence of ESBL genes have also been reported from different locations in three dairy farms stating the possibility for intramammary infections in cows [28].
Figure 2: PCR amplified fragments blaTEM (on left of the ladder); Positive control in-house control (E. coli; Resistant to Aztreonam, Cefoxitin, ampicillin, Cefotaxime, Ceftazidime, Phenotype confirmed), Negative control - E. coli ATCC 25922, 1-3 - bacterial isolates (E. coli), and blaSHV (on right of the ladder); Positive control - K. pneumonia ATCC 700603, Negative control - E. coli ATCC 25922, 1-4 bacterial isolates (E. coli) on agarose gel (1.5%).
Figure 3: PCR amplified fragments of blaAmpC on 1.5% agarose gel; Positive control - in-house control (E. coli; Resistant to Aztreonam, Cefoxitin, ampicillin, Cefotaxime, Ceftazidime), Negative control - autoclaved distilled water, Samples 1 to 10 - bacterial isolates of E. coli.
Figure 4: PCR amplified fragments on 1.5% agarose gel of blaCTX-M; Positive control - in-house control (E. coli; Resistant to Aztreonam, Cefoxitin, ampicillin, Cefotaxime, Ceftazidime), Negative control - autoclaved distilled water, Samples 1 to 3 - bacterial isolates of E. coli.
Figure 5: Bar diagram representing the percentage of ESBL gene detected in the E. coli isolates.
Blagenes
Percentage
(no. of isolates)
SHV + TEM
21.42(6)
SHV + CTX-M
10.71(3)
TEM + CTX-M
28.57(8)
TEM + AmpC
85.71(24)
SHV+ TEM + CTX-M
10.71(3)
Table 1: Table representing presence of two or more bla genes in the E. coli isolates from cow dung (n=28).
Cow dung is used as an organic fertilizer for the cultivation of food crops in agricultural field, on a large scale as well as in kitchen gardens. The occurrence of ESBL producing enteric E. coli in the fecal microflora of cows (in our study) represents an obvious risk for contamination of raw food products and also that of animal origin furthering the dissemination of ESBL producing isolates into the environment. More recently, reports have also raised concern about the dissemination of ESBL producing E. coli in healthy food producing animals in several countries. Therefore, the impact of healthy farm animals as a possible reservoir for ESBL producing Enterobacteriaceae on the food processing chain need to be assessed. Given the relatively high occurrence of ESBL producers in faecal samples from cows in our study, it is striking, that the rate of ESBL producers has alarmingly increased over the last few years.
Both methanol and aqueous extracts of Woodfordia fructicosa (L.) Kurz flowers showed a zone of inhibition against TEM + SHV positive isolates in comparison to Holarhenna antidysenterica Wall. (Table 2) with comparable inhibitory zones between both the plant samples. The results observed from the screening of plant specimens for their antibacterial efficacy indicates the need for the assessment of medicinal plants against ESBL producing isolates and the possibility of finding newer antibacterial agents to tackle the emerging antimicrobial resistance in Enterobacteriaceae. Considering the rich diversity of plants, it is expected that screening and scientific evaluation of plant extracts for their anti-microbial activity may provide new anti-microbial substances. Hence the present investigation clearly reveals the antibacterial nature of the medicinal plants and suggests that the plants could be exploited in the management of diseases caused by these bacteria in both human and animal systems. In this study, we aimed to provide an informative basis on the presence of ESBLs in cows in Assam, India and the possibility of exploring newer antimicrobial agents from plant samples indigenous to North EasternIndia and Assam.
Plant
Parts used
Solvent
Concentration (mg/mL)
Average Zone of Inhibition (mm)*
Holarrhena antidysenterica Wall
Leaves
Methanol
400
11.0
Aqueous
300
-
Woodfordia fructicosa (L.) Kurz
Flowers
Methanol
400
10.3
Aqueous
400
10.3
Table 2: Table representing diameter of the zone of inhibition (in mm) of the medicinal plants tested against three E. coli isolates possessing TEM and SHV genotype.
ESBLs are the main cause of resistance to beta-lactam antibiotics in E. coli. Due to the clinical importance of the detection of ESBLs, screening and confirmatory methods have been routinely used to investigate the production of these enzymes in E. coli species. As their occurrence has been increasing, it becomes essential to evaluate their occurrence in this population. Expanded spectrum cephalosporins are β-lactams with a broad spectrum of activity against most Gram negative bacteria. However they are sensitive to hydrolysis by ESBLs. The dissemination of these enzymes is currently a global problem [29,30]. It is also well known that plasmids carrying genes encoding ESBLs may also carry genes encoding resistance to non β-lactam antibiotics such as aminoglycosides, chloramphenicol and trimethoprim-sulphamethoxazole [31]. Relatively very little is known about the epidemiology of ESBLs in veterinary medicine. There have been a few studies reporting ESBLs from farm animals and pets [32- 34]. In fact plasmid encoded ESBLs which are once rarely detected of animal origin bacteria are more frequently being observed in therecent years. The factors leading to the emergence of ESBLs among bacteria of animal origin are not fully elucidated.
Conclusion
To our knowledge, this is the first report describing genetic basis of ESBLs in Enterobacteriaceae detected from food animals in Assam, India. Larger scale surveillance studies in veterinary are needed to track the evolution and epidemiology of ESBLs. Development of newer antibiotic candidates for dealing with the ever emerging bacterial resistance mechanisms of ESBL positive organisms has become the need of the hour. Moreover, strict drug policies need to be implemented to limit the irrational use of antibiotics. Surveillance studies need to be done from time to time to keep track of the extent of resistance acquired by the organisms in a particular area and also to identify any new mechanisms developed, if any.
References
- Alanis AJ. Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res. 2005; 36: 697-705.
- Hawkey PM, Jones AM. The changing epidemiology of resistance. J Antimicrob Chemother. 2009; 64: 3-10.
- Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011; 24: 718-733.
- Ewers C, Antão EM, Diehl I, Philipp HC, Wieler LH. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl Environ Microbiol. 2009; 75: 184-192.
- Jong AD, Bywater R, Butty P, Deroover E, Godinho K, Klein U, et al. A pan-European survey of antimicrobial susceptibility towards human-use antimicrobial drugs among zoonotic and commensal enteric bacteria isolated from healthy food-producing animals. J Antimicrob Chemother. 2009; 63:733-744.
- O'Keefe A, Hutton TA, Schifferli DM, Rankin SC. First detection of CTX-M and SHV extended-spectrum beta-lactamases in Escherichia coli urinary tract isolates from dogs and cats in the United States. Antimicrob Agents Chemother. 2010; 54: 3489-3492.
- Hawkey PM. The growing burden of antimicrobial resistance. J Antimicrob Chemother. 2008; 62: 1-9.
- Knothe A, Shah P, Kremery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime and in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983; 11: 315-317.
- Philippon A, Labia R, Jacoby G. Extended-spectrum beta-lactamases. Antimicrob Agents Chemother. 1989; 33: 1131-1136.
- ESBLs.UK. A threat to human and animal health? -A joint report by the Joint working group of DARC and ARHAI. 2012.
- Bhattarai S, Bhuju DR. Antimicrobial activity of useful parts of Woodfordia fructicosa (Linn.) Kurz. of Nepal. Inter J Pharma Biol Archives. 2011; 2: 756-761.
- Mahato S, Mehta A, Roy S. Studies on antibacterial effects of bark, seed and callus extracts of Holarrhena antidysenterica Wall. The Bioscan (Supplement on Medicinal plants). 2013; 8: 717-721.
- Ganapathy PSS, Ramachandra YL, Sudeep HV, Bellamakondi PK, Achar KGS, Rai SP. Pharmacognostic and phytochemical evaluation of Holarrhena antidysenterica Wall. Asian & Australasian J Plant Sci Biotech. 2009; 3: 47-50.
- Pandey P, Mehta A, Hajra S. Evaluation of Antimicrobial Activity of Ruta graveolens Stem extracts by Disc Diffusion Method. Journal of Phytology. 2011; 3: 92-95.
- Levine M, Epstein SS, Vaughn RH. Differential Reactions in the Colon Group of Bacteria. Am J Public Health Nations Health. 1934; 24: 505-510.
- Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved Standard-Second Edition. NCCLS document M31-A2, 2002.
- Lal P, Kapil A, Das BK, Sood S. Occurrence of TEM & SHV gene in extended spectrum beta-lactamases (ESBLs) producing Klebsiella sp. isolated from a tertiary care hospital. Indian J Med Res. 2007; 125: 173-178.
- Jemima SA, Verghese S. Multiplex PCR for bla(CTX-M) & bla(SHV) in the extended spectrum beta lactamase (ESBL) producing gram-negative isolates. Indian J Med Res. 2008; 128: 313-317.
- Briñas L, Zarazaga M, Sáenz Y, Ruiz-Larrea F, Torres C. Beta-lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrob Agents Chemother. 2002; 46: 3156-3163.
- Petchi RR, Vijaya C, Parasuraman S. Anti-arthritic activity of ethanolic extract of Tridax procumbens (Linn.) in Sprague Dawley rats. Pharmacognosy Res. 2013; 5: 113-117.
- Ikpefan EO, Fajana A, Olowojoba J. Cytotoxic and Growth Inhibitory Effects of the Methanol Extract of Tridax procumbens Linn (Asteraceae). J Pharmacogn Phytochem. 2013; 2: 26-31.
- Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, et al. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg Infect Dis. 2012; 18: 741-749.
- Madec JY, Lazizzera C, Châtre P, Meunier D. Prevalence of Fecal Carriage of Acquired Expanded-Spectrum Cephalosporin Resistance in Enterobacteriaceae Strains from Cattle in France. J Clin Microbiol. 2008; 46:1566-1567.
- Kucukbasmaci O, Ciftcioglu G, Midilli K, Issa G. Detection of extended spectrum β-Lactamase producing Enterobacteriaceae from food animals In Turkey. Revue de Médecine Vétérinaire. 2008; 159: 586-592.
- Duan RS, Sit TH, Wong SS, Wong RC, Chow KH, Mak GC, et al. Escherichia coli producing CTX-M beta-lactamases in food animals in Hong Kong. Microb Drug Resist. 2006; 12: 145-148.
- Shiraki Y, Shibata N, Doi Y, Arakawa Y. Escherichia coli producing CTX-M-2 beta-lactamase in cattle, Japan. Emerg Infect Dis. 2004; 10: 69-75.
- Geser N, Stephan R, Hachler H. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Veterinary Research. 2012; 8: 1-9.
- Nobrega DB, Guiduce MVS, Guimaraes FF, Riboli DF, Cunha MLRS, Langoni H, et al. Molecular epidemiology and extended spectrum β-lactamases production of Klebsiella pneumonia isolated from three dairy herds. Pesq Vet Bras. 2013; 33: 855-859.
- Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005; 18: 657-686.
- Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol. 2007; 7: 459-469.
- Paterson DL, Mulazimoglu L, Casellas JM, Ko WC, Goossens H, Von Gottberg A, et al. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum beta-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin Infect Dis. 2000; 30: 473-478.
- Costa D, Poeta P, Briñas L, Sáenz Y, Rodrigues J, Torres C. Detection of CTX-M-1 and TEM-52 beta-lactamases in Escherichia coli strains from healthy pets in Portugal. J Antimicrob Chemother. 2004; 54: 960-961.
- Briñas L, Moreno MA, Teshager T, Sáenz Y, Porrero MC, Domínguez L, et al. Monitoring and characterization of extended-spectrum beta-lactamases in Escherichia coli strains from healthy and sick animals in Spain in 2003. Antimicrob Agents Chemother. 2005; 49: 1262-1264.
- Liebana E, Batchelor M, Hopkins KL, Clifton-Hadley FA, Teale CJ, Foster A, et al. Longitudinal farm study of extended-spectrum beta-lactamase-mediated resistance. J Clin Microbiol. 2006; 44: 1630-1634.