Research Article
Austin J Biotechnol Bioeng. 2014;1(1): 7.
Novel Approach towards Synthesis of Silver Nanoparticles from Myxococcus virescens and their Lethality on Pathogenic Bacterial Cells
Wioletta Wrótniak -Drzewiecka1, Swapnil Gaikwad2, Dariusz Laskowski1, Hanna Dahm1, Janusz Niedojadlo3, Aniket Gade2,4 and Mahendra Rai1,2*
1Department of Microbiology, Nicolaus Copernicus University, Lwowska, Poland
2Department of Biotechnology, Sant Gadge Baba Amravati University, India
3Department of Cell Biology, Nicolaus Copernicus University, Poland
4Department of Biology, Utah State University, USA
*Corresponding author: Mahendra Rai K, Department of Biotechnology, Sant Gadge Baba Amravati University, Amravati- 444602 Maharashtra, India.
Received: June 30, 2014; Accepted: July 25, 2014; Published: July 28, 2014
Abstract
Aim: We report extracellular synthesis of silver nanoparticles by cell filtrate of Myxococcus virescens. Moreover, the effect of silver nanoparticles on survivability or lethality of human pathogenic bacteria viz. Staphylococcus aureus (ATCC 6538), Bacillus subtilis subsp. spizizenii (ATCC 6633) and Pseudomonas aeruginosa (ATCC 10145) has been studied.
Methods and Results: The silver nanoparticles were synthesized from M. virescens cell filtrate, and characterized by UV-Vis Spectroscopy, Nanoparticle Tracking Analysis (NTA) by LM 20, Zeta Potential analysis, Fourier Transform Infra-Red Spectroscopy (FTIR) and Transmission Electron Microscopy (TEM). Antibacterial efficacy of silver nanoparticles was tested which comprised the effect of survivability or lethality of human pathogenic bacteria. Biosynthesized silver nanoparticles demonstrated promising cell lethality for human pathogenic bacteria.
Conclusion: M. virescens mediated synthesized silver nanoparticles are quite stable, and capped with proteins. This is a sustainable, eco-friendly and simple process for synthesis of desirable silver nanoparticles which is havinglethal properties against some clinical bacterial cell.
Significance and Impact of Study: M. virescens mediated synthesized nanoparticles could make a starting point to obtain new antibacterial substances towards multiple antibiotic-resistant pathogenic bacteria.
Keywords: Myxococcus virescens, Silver Nanoparticles, Human pathogens, Cell lethality, Antibacterial activity
Introduction
Nanobiotechnology is amalgamation between nanotechnology and biology, which refers to the ability to create and manipulate biological and biochemical materials, devices, and systems at nano level [1]. In nanotechnology, for development of new nanostructure materials and devices it requires nanoparticles with unique properties. Silver has been used widely since ancient time to treat infection and prevent spoilage [2]. When the era of antibiotic began the use of silver for its antimicrobial properties decreased [3]. Since antibiotic resistant microorganisms emerged the interest to use silver as antimicrobial agent is rising again. Zero valent silver (Ag0) nanoparticles are a valuable alternative for ionic silver. Due to their large specific surface to volume ratio nanoparticles have different properties than ionic silver. Silver nanoparticles are active against broad spectrum Gram-negative and Gram-positive bacteria. Furthermore, silver nanoparticles showed antifungal [4] and antiviral activity [5].
Biosynthesis of silver nanoparticles is an environmental-friendly method without the use of toxic and expensive chemicals. The ability of microorganisms in production of metal nanoparticles has opened a new exciting approach toward the development of these natural nano-factories [6]. The important aspect in the process of producing highly stable, well-characterized and and highly active nanoparticles are selection of the best organisms and optimization of conditions for growth and enzyme activity. These factors could control morphology, size, stability, aggregation and other properties of bionanoparticles. Microorganisms demonstrated more advantages over other biological systems because of their high tolerance towards the heavy metals [7].
Due to physicochemical properties, silver nanoparticles have been extensively utilized and are currently used as antibacterial agents in fruit storage [8], textile and health industries [9], also for labeling and as biosensors [10]. To achieve the nanoparticles with their unique properties, there is a pressing need to develop a cheaper and eco-friendly method for synthesis of nanoparticles which eliminates the use of toxic chemicals during their synthesis process. Metal nanoparticles like silver, gold and platinum are extensively exploited for the welfare of human beings [11]. The synthesis of silver nanoparticles is broadly studied by chemical and physical methods [12]. Biological methods using plants [13,14], fungi [15], and bacteria [16] have superior option for chemical and physical synthesis of silver nanoparticles.
In recent times microorganisms and plants are found as environment-friendly nanofactories. A wide range of microbes including bacteria like, Pseudomonas aeruginosa [16], Bacillus sp. [17], Bacillus cereus [18], cyanobacteria-Spirulina platensis [19], actinobacteria-Streptomyces sps. [20], Bacillus subtilis [21] and fungi like Fusarium oxysporum [22], Fusarium acuminatum [15], Fusarium solani [23], Phoma glomerata [24], Alternaria alternata [25], Fusarium culmorum [26], Neurospora crassa [27], Trichoderma [28], Fusarium graminearum, Fusarium scirpi [29] were exploited for the synthesis of silver nanoparticles. Plants like Gliricidia sepium (Jacq.) [30], Carica papaya [31], Opuntia ficus-indica [13], Allium cepa [32], Weed [33], Foeniculum vulgare [34], Ocimum sanctum [35], Cassia auriculata [36], Phyllostachys sp. [37], Soap nuts [38], Murraya koenigii [39], Lawsonia inermis [40] were also found to be important source for green synthesis of silver nanoparticles. Microorganisms demonstrated more advantages over other biological systems because of their high tolerance towards the heavy metals like Fe, Co, Ni, Cu, and Zn, As, Cd, Hg, Pb or U [41-43].
Myxobacteria (slime bacteria) are included among the Delta group of Proteobacteria. They are found in the topsoil where they feed on organic matter and prey on other microorganisms by secreting hydrolytic enzymes and antimicrobials [44]. Antibiotic and enzymes produced by myxobacteria kill microorganisms and lyse cells. Reduction of silver ions is catalyzed by enzymes or by nonenzymatic interaction of Ag+ with cell wall functional groups. In the current studies we focused on synthesis of silver nanoparticles from cell supernatant of M. virescens and the influence of silver nanoparticles on the growth of human pathogenic bacteria.
Materials and Methods
Test myxobacterium
Myxococcus virescens was isolated from top level of the forest soil (depth 10 cm) under Scots pine (Pinus sylvestris L.) trees of Torun, Poland. The chemical properties of the soil were as follows: pH H2O- 4.22, pH KCl-3.44, C org. % 1.78, and N % 0,075, C/N24. Soil samples were moisturized using water with dissolved antibiotics: actidione (100 mg/dm3), nystatine (100 mg/dm3) and penicillin (50 mg/dm3). For inoculation of myxobacteria, lumps of soil (weighing ca. 0.1 g) as a inoculums were used, which were put onto petri dishes containing solidified Vy/2 myxobacteria-semi selective medium (Baker's yeast 0.5%, CaCl22H2O 0.1%, cyanocobalamin 0.5ug ml-1, agar 1.5%, pH7.2) (Reichenbach and Dworkin 1981). Inoculated plates were incubated at 30°C for 30 days. Myxobacteria were subcultured and stored on Vy/2 agar medium.
Preliminary identification of myxobacterial strain was carried out on the basis of diagnostic keys by Reichenbach and Dworkin M [45]. The strain was identified by 16s RNA analysis. Genomic DNA was extracted and amplified following the procedures given by [46]. The isolated DNA was amplified by PCR technique, using the following primers: 10-30F and 150°R. The samples were subjected to the preliminary denaturation (98°C, 3 min.) in the thermocycler. Amplification (28 cycles) was carried out using the following thermalprofile: 93°C for 1 minute (denaturation), 52°C for 1 minute (joining the starters), and 72°C for 2 minutes (elongation of the starters). The final elongation of the starters was carried out at temperature 52OC (1 min.) and 72°C (10 min.). To obtain the specific products of PCR (size - 1584bp), Q/Aquick Gel Extraction Kit (Qiagen) was used. The length of amplified fragment of DNA was estimated on the basis of molecular weight standard. In our studies, we used DNA Marker III (Roche) containing DNA fragments of length from 564 to 21226bp. The replicated and purified genetic material was sequenced using ABI Prism TM Big Bye TM Terminator Cycle Sequencing Kit. The sequence consensus was compared with NCBI Gene Bank data base.
Extracellular synthesis of silver nanoparticles
For the synthesis of silver nanoparticles, 100 ml CAS broth [47] was prepared in flask, sterilized and inoculated with fresh growth of Myxococcus virescens. The inoculated flasks were incubated at 30°C for 72 hours. The culture was then centrifuged at 12,000 rpm for 15 minutes and supernatant was discarded. Bacterial cells settled at bottom were washed 2-3 by sterile distilled water to remove media traces. Afterwards, bacterial cells were suspended in sterile distilled water and incubated for 48 hours. After incubation, suspension was centrifuged at 12,000 rpm and cell free filtrate which contains osmotically lysed bacterial cell content was treated with 1mM AgNO3 and incubated for 48 hours. Only cell filtrate and broth with 1mM AgNO3 were also maintained as control. The experiment was set up in triplicate.
Detection of silver nanoparticles
After the 48 hr incubation of cell filtrate and AgNO3 (mixture), the preliminary detection of silver nanoparticles was carried out by visual observation of colour change in filtrate. All the above reaction mixtures were then subjected to UV-visible Spectrophotometer analysis (Shimadzu UV-1700, Japan). The spectrum was scannedfrom 200-800 wavelengths at 1 nm resolution.
Antibiotic susceptibility profile
Nanoparticle tracking analysis (NTA) by LM 20
Liquid samples of silver nanoparticles were used to perform NTA analysis, in which laser beam (approximately 40mW at k=635 nm) was passed through a scattering cell. Particles present within the path of the laser beam were observed via a dedicated non-microscope optical instrument (LM-20, NanoSight Pvt. Ltd., UK) having Charge Coupled Device (CCD) camera. The motion of the particles in the field of view (approximately 100×100 μm) was recorded (at 30 fps) and the subsequent video and images were analyzed for the size distribution of nanoparticles in sample.
Measurement of zeta potential
The zeta potential was measured by using a Zetasizer Nano ZS 90 (Malvern Instrument ltd, UK). Measurements were made by means of Dynamic Light Scattering (DLS) in the range of 0.1-1000 μm. 1000 μl of sample was transferred in the clear disposable zeta cell for the measurement of zeta potential. The zeta potential was calculated using Henry's equation.
Fourier transforms infra-red (FTIR) spectroscopy
Silver nanoparticles were further characterized by FTIR (Perkin- Elmer FTIR- 1600, USA) in the range 500-2000 cm-1 at a resolution of 4 cm-1 to understand the biomolecules responsible for the reduction of silver ions and stabilization of silver nanoparticles in the mixture. For sample preparation, 300 μl of concentrate colloidal silver nanoparticle solution was mixed with 10 mg potassium bromide (KBr) in clean crucible, until it becomes a fine powder. The sample was prepared and oven dried to remove the traces of moisture.
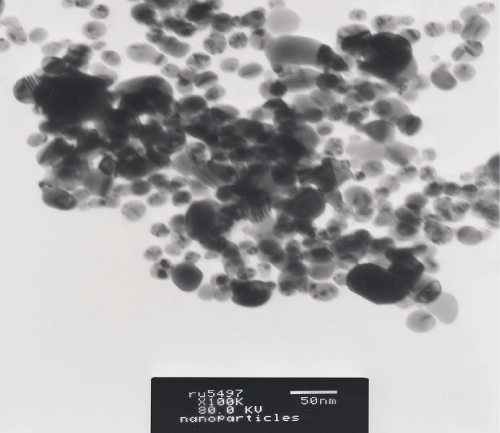
Transmission Electron Microscopic Analysis
To determine the size and shape of silver nanoparticles Transmission Electron Microscopic analysis (TEM) was carried out. The silver nanoparticles were characterized by TEM (Joel 1010, at 80kv), on conventional carbon coated copper grids (400 meshes,), it was cleaned using plasma treatment under oxygen for 45 sec. A 5 μl of sample was then placed on the grid and dried at room temperature for 1 hour. The samples were inspected by operating at 80 KV. Images ofsample were taken to have a clear representation of its composition.
Removal of unreacted Ag+
To remove the unreacted Ag+ from nanoparticle sample, it was treated with NaCl solution. After addition of NaCl, Ag+ reacts with Cl- and form white precipitate of AgCl. The precipitate was then removed by centrifugation of mixture at 4,000 rpm for 15 minutes. After removing the pellet, the supernatant was centrifuged at 14,000 rpm for 30 min to concentrate the silver nanoparticles, and then dried at 50°C. Subsequently, their dry mass was estimated. To be used in further studies silver bionanoparticles were resuspended in deionized water, to obtain their desired concentration [23].
Analysis of antibacterial activity of silver nanoparticles
Antibacterial efficacy of silver nanoparticles was tested against: Staphylococcus aureus ATCC 6538, Bacillus subtilis subsp. spizizenii ATCC 6633 and Pseudomonas aeruginosa ATCC 10145.
The studies comprised: (a) effect of biosynthesized silver nanoparticles on the bacterial growth, using double-layer agar plates, b) the effect of biosynthesized silver nanoparticles on survivability or lethality of bacteria.
a) Effect of biosynthesized silver nanoparticles on the bacterial growth, using double-layer agar plates [48].
Molten and cooled to 45°C Trypticase Soy Agar (TSA) medium in culture tube was inoculated with the cells suspension 104 cfu ml-1 of the respective bacteria (obtaining a final concentration of bacterial cells 103cfu ml-1) and supplemented with silver nanoparticles up to a concentration 100 μg ml-1. Subsequently, after mixing, medium was poured as a second layer onto an already prepared petri dish of TSA agar and set aside to solidify. Plates with the poured medium without nanoparticles were also kept as a control. Plates were incubated for 24 hrs at 37°C. Bacterial colonies grown on the plates were counted. Anextent of antibacterial activity of silver nanoparticles was calculated by following formula:
AE (%) = [(C - T)/C] * 100
where
AE: Antibacterial activity (expressed in percents),
C: Number of bacterial colonies on the control plate (cfu/ per sample),
T: Number of bacterial colonies on the experimental plate (with silver nanoparticles) (cfu/ per sample).
b) The effect of biosynthesized silver nanoparticles on survivability or lethality of bacteria
LIVE/DEAD® BacLight™ Bacterial Viability Kit, L7012 (Invitrogen) stain was used, which makes possible a discrimination between live bacterial cells (green fluorescence) and the dead ones (red fluorescence) under the fluorescence microscope. Respective bacterial cultures were grown in liquid TSB medium for 24 hours at 37°C, and after that it was centrifuged at 5000 rpm for 5 min at 5°C. The bacterial pellet was then rinsed with sterile physiological saline, suspended in physiological saline maintained to the optical density equal to 4 McFarland units. Thus obtained cell suspension was 10- fold diluted by addition of the appropriate volume of nanoparticles suspension or silver ions solution used for comparison to obtain the final concentration equal to 75 μg ml-1. Silver nanoparticles were purified according to the procedure described by [23]. Bacterial suspensions without silver nanoparticles were used as controls. Samples were incubated for 30 min. at room temperature, and then rinsed with physiological saline and resuspended in sterile distilled water. After staining with LIVE/DEAD stain, the bacterial suspension was put onto glass microscopic slide and was observedunder the epifluorescence microscope ZEISS Axiostar plus, with the light source mbq 52 ac (Carl Zeiss, Jena, Germany), using the oil immersion 100-fold magnifying objective. Live and dead bacterial cells on microphotographs were counted with computer aid, using image analysis software program MacBiophotonics Image J (version 1.41a) and its Cell Counter plug-in.
Results
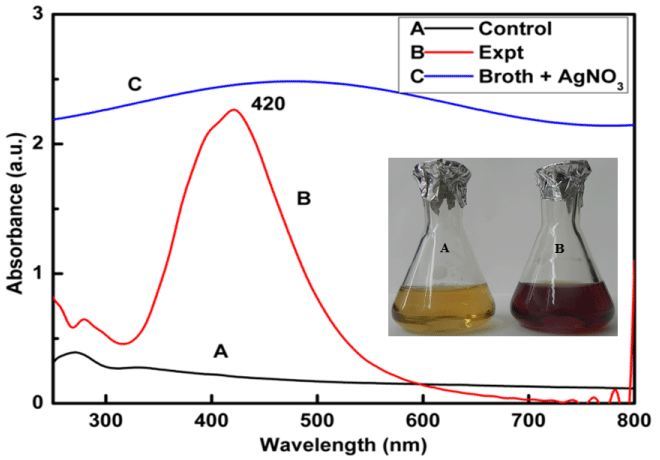
Synthesis of silver nanoparticles using Myxococcus virescens was found to be a promising method because change in colour of cell filtrate was observed from yellowish or colourless to brown when treated with aqueous 1mM AgNO3 (Figure 1 inset). Synthesized silver nanoparticles were then subjected to UV-Visible spectroscopy showing the absorbance peak at 420 nm (Figure 1), which is characteristic feature of silver nanoparticles.
Figure 1: UV-Visible Spectra of (Control, Spectra A) cell free extract of Myxococcus virescens, silver Nanoparticles (Expt., Spectra B) synthesized by cell free extract of Myxococcus virescens, and broth + 1mM AgNO3 (Spectra C). Inset figure: colour change of bacterial filtrate from pale yellow (control) (A) to dark brown (treated) after synthesis of Silver Nanoparticles (B).
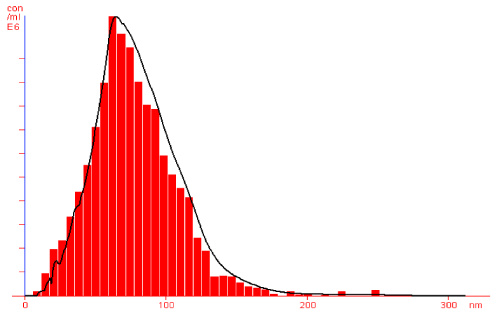
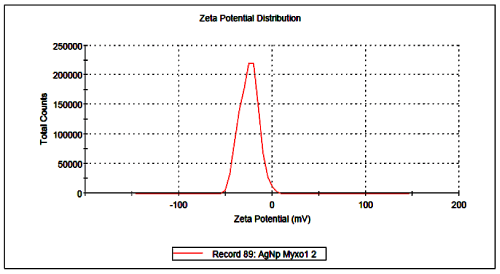
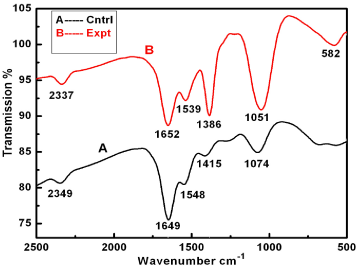
Further, silver nanoparticles were characterized by Nanoparticle Tracking Analysis (NTA) using Nanosight LM 20 to analyse size and size distribution of particles on the basis of their brownian motion in suspension. The average size of silver nanoparticles synthesized by M. virescens was 81 nm and mode size 65 nm, means in suspension maximum nanoparticles are of 65 nm size (Figure 2). In addition, to determine the stability of synthesized silver nanoparticles, zeta potential was measured. Figure 3 reveals that the zeta potential of synthesized silver nanoparticles was -25.2 mV, which indicates that the synthesized nanoparticles were moderately stable. Further analysis was performed by FTIR spectroscopy to acquire the idearegarding the protein capping and interactions of proteins with silver, which might be responsible for synthesis and stabilization of silver nanoparticles. In FTIR spectrum (Figure 4) of control (Figure 4 spectra A) peaks were observed at 1074, 1415, 1548, 1649 and 2349cm-1. After the synthesis of silver nanoparticles these peaks were shifted to 1051, 1386, 1539, 1652 cm-1 respectively. Transmission Electron Microscopy (TEM) revealed (Figure 5) that the synthesized silver nanoparticles were polydispersed and spherical in shape with the size distribution 7- 50 nm.
Figure 2: NTA (Nanosight-LM 20) nanoparticle size distribution histograms showing Polydisperse silver nanoparticles synthesized by cell free extract of Myxococcus virescens.
Figure 3: Surface zeta potential graph showing negative zeta potential value for silver nanoparticles synthesized by cell free extract of Myxococcus virescens.
Figure 4: FTIR spectra of bacterial cell filtrate (control, A), and silver nanoparticles (experimental, B) synthesized by cell free extract of Myxococcus virescens.
Figure 5: TEM micrograph showing spherical and polydispers silver nanoparticles synthesized by cell free extract of Myxococcus virescens.
Our study on, the effect of silver nanoparticles on the growth of bacteria with the use of double-layer plate method have shown that the concentration of nanoparticles used (100 μg ml-1) exerted an antibacterial action, which was within the range 25.8-33.9%. The highest inhibitory effect was observed in the Gram-negative bacterium; Pseudomonas aeruginosa (Table 1).
Bacterial species
Number of bacterial colonies on plate (cfu) x103 (control)
Number of bacterial colonies (cfu) x 103 grown on plate in the presence of silver nanoparticles (AgNPs)
AE [%] (percentage antibacterial activity)
100 μg cm-3 of AgNPs
B.subtilis subsp. spizizenii
252
187
25.80
S. aureus
510
382
25.10
P. aeruginosa
784
518
33.93
Table 1: Effect of silver nanoparticles synthesized by cell free extract of Myxococcus virescens on bacterial growth on agar medium.
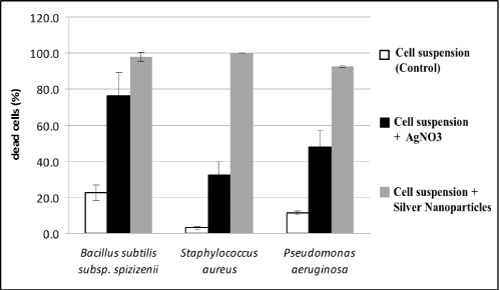
Results of our studies on the effect of silver nanoparticles or silver ions on the lethality of bacterial cells, estimated that lethality of all the strains studied was higher in the presence of silver nanoparticles (over 90%) as compared to silver nitrate as a control. The strongest action of silver nanoparticles was noted for Staphylococcus aureus (Figure 6). However, both in case of test dealing with the effect of silver nanoparticles on bacterial growth, and their effect bacterial survival, antimicrobial activity of nanoparticles studied essentially did not differ depending on the strain.
Figure 6: Effect of silver nanoparticles synthesized by cell free extract of Myxococcus virescens and silver ions on the lethality of pathogenic bacteria cells (BacLight method) (average values ± standard deviation).
Discussion
The change in colour from pale yellow to dark brown was due to the reduction of silver ions (Ag+) to silver nanoparticles (Ag0) which is the evidence for synthesis of silver nanoparticles (Figure 1 inset). Silver nanoparticles exhibit strong absorbance at 420 nm in the visible range due to surface plasmon resonance [49]. The single and symmetric peak indicates the synthesis of spherical nanoparticles. synthesis of spherical nanoparticles. These findings support the earlier studies carried out by many researchers using different bacteria and fungi as biological agents for the synthesis of silver nanoparticles [15,2,26,50,16,18,51,52].
Negative zeta potential value of the particles might be due to adsorption of OH- ions on it. Since adsorption of OH- ions on the silver nanoparticles increase zeta value of the nanoparticles, there is an increase in stability of the nanoparticles due to electrostatic repulsion among the negative charges. An OH- ion helps in preventing the aggregate formation and maintains the smaller size of silver nanoparticles [53]. In FTIR analysis of silver nanoparticles peak observed at 1051cm-1 associated with stretch vibration of -C-Obond.The peak at 1386 cm-1 is due to NO-3 existed in the residual solution [54]. The band at 1539 and 1652 cm-1 arose due to N-O nitro group and Stretch vibration of -C=C- respectively. These results corroborate with findings of Haung et al. [54]. As shown in Figure 4 spectra B, the peak exhibits the binding of amide linkage with silvernanoparticles which was clearly indicated in the infrared region of the electromagnetic spectrum indicating the presence of protein as a capping agent for silver nanoparticles. Proteins have stronger ability to bind silver nanoparticles which increases the stability of synthesized nanoparticles [55].
Raheman et al. [50] observed greater antibacterial effect of silver nanoparticles in case of Staphylococcus aureus than in Gram negative bacteria Salmonella typhi., Sunkar and Nachiyar [18] also demonstrated bactericidal activity of silver nanoparticles to pathogenic strains of bacteria namely Gram negative (Escherichia coli, Pseudomonas aeruginosa) and Gram positive bacteria (Staphylococcus aureus). Effect silver nanoparticle was observed to be more profound in Gram negative bacteria than Gram positive. Authors suggested that it is attributed to the fact that the relative abundance of negative charges on Gram negative bacteria facilitated the interaction between the nanoparticles and the cell wall.
The bactericidal effect of silver nanoparticles has been well established; however, the mechanism is only partially understood. The silver nanoparticles show efficient antimicrobial property compared to other salts due to their extremely large surface area, which provides better contact with microorganisms. The nanoparticles get attached to the cell membrane and also penetrate inside the bacteria. The silver nanoparticles are considered to be a slow release source of silver ions, which interact with sulfur-containing proteins present in bacterial cell membrane as well as with the phosphorus containing compounds like DNA [56,57]. When silver nanoparticles enter in the bacterial cell it forms a low molecular weight region in the center of the bacteria due to which the bacteria conglomerates and protect the DNA from the silver ions. The nanoparticles preferably attack the respiratory chain, cell division finally leading to cell death. Moreover, the nanoparticles release silver ions in the bacterial cells, which enhance their bactericidal activity [2].
Extracellular synthesis of silver nanoparticles by Myxococcus virescens is being reported for the first time. It has been confirmed that cell filtrate of M. virescens is capable of synthesizing the silver nanoparticles. M. virescens mediated synthesized silver nanoparticles are quite stable, and capped with proteins. This is a sustainable, eco-friendly and simple process for synthesis of desirable silver nanoparticles. Biosynthesized silver nanoparticles reveal antibacterial properties against some clinical bacteria showing lethal effect on their cells. These nanoparticles could make a starting point to obtain new antibacterial substances towards multiple antibiotic-resistant pathogenic bacteria.
Acknowledgement
This research has been financed by a grant from Polish Ministry of Science and Higher Education (Grant No. NN309427438). We also thank UGC New Delhi for support under UGC-SAP programme.
References
- Kholoud MM, El-Nour A, Eftaiha A, Abdulrhman AQ, Ammar AA. Synthesis and applications of silver nanoparticles. Arabian J. Chem. 2010; 3: 135-140.
- Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009; 27: 76-83.
- Sintubin L, De Windt W, Dick J, Mast J, van der Ha D, Verstraete W, et al. Lactic acid bacteria as reducing and capping agent for the fast and efficient production of silver nanoparticles. Appl Microbiol Biotechnol. 2009; 84: 741-749.
- Kim KJ, Sung WS, Moon SK, Choi JS, Kim JG, Lee DG, et al. Antifungal effect of silver nanoparticles on dermatophytes. J Microbiol Biotechnol. 2008; 18: 1482-1484.
- Gaikwad SC, Ingle AP, Gade AK., Rai MK, Falanga A, Incoronato N, et al. Antiviral activity of mycosynthesized silver nanoparticles against Herpes Simplex virus and Human Parainfluenza Virus Type 3. Int. J Nanomed. 2013; 8: 4303-4314.
- Iravani S. Green synthesis of metal nanoparticles using plants. Green Chem. 2011; 13: 2638-2650.
- Durán N, Marcato PD, Durán M, Yadav A, Gade A, Rai M, et al. Mechanistic aspects in the biogenic synthesis of extracellular metal nanoparticles by peptides, bacteria, fungi, and plants. Appl Microbiol Biotechnol. 2011; 90: 1609-1624.
- Gudadhe J, Yadav A, Gade A, Marcato PD, Duran N, Rai M. Preparation of an agar-silver nanoparticles (A-AgNp) film for increasing the shelf-life of fruits, IET Nanobiotechnol. 2013.
- Durán N, Marcato PD, Souza GIH De, Alves OL, Esposito E. Antibacterial Effect of Silver Nanoparticles Produced by Fungal Process on Textile Fabrics and Their Effluent Treatment. J. Biomed. Nanotechnol. 2007; 3: 203-208.
- Rai M, Gade A, Yadaw A. Biogenic nanoparticles: an introduction to what they are, how they are synthesized and their applications. In: Metal Nanoparticles in Microbiology. Rai M, Duran N, Editors. 2011. 1-14.
- Song JY, Kim BS. Rapid biological synthesis of silver nanoparticles using plant leaf extracts. Bioprocess Biosys Eng. 2008; 32: 79-84.
- Natarajan K, Selvaraj S, Ramachandra Murty V. Microbial Production of Silver Nanoparticles Digest J. Nanomater. & Biostruct. 2010; 5: 135 -140.
- Gade AK, Gaikwad SC, Tiwari V, Yadav A, Ingle AP, Rai MK. Biofabrication of silver nanoparticles by Opuntia ficus-indica: In vitro antibacterial activity and study of the mechanism involved in the synthesis. Curr. Nanosci. 2010; 6: 370-375.
- Mittal AK, Chisti Y, Banerjee UC. Synthesis of metallic nanoparticles using plant extracts. Biotechnol. Adv. 2013; 31: 346-356.
- Ingle A, Gade A, Pierrat S, Sonnichsen C, Rai M. Mycosynthesis of silver nanoparticles using the fungus Fusarium acuminatum and its activity against some human pathogenic bacteria. Curr Nanosci. 2008; 4: 141-144.
- Deshmukh SD, Deshmukh SD, Gade AK, Rai MK. Pseudomonas aeruginosa Mediated Synthesis of Silver Nanoparticles Having Significant Antimycotic Potential Against Plant Pathogenic Fungi. J. Bionanosci. 2012; 6: 90-94.
- Pugazhenthiran N, Anandan S, Kathiravan G, Kannaian N, Prakash U, Crawford S, et al. Microbial synthesis of silver nanoparticles by Bacillus sp. J. Nanopart. Res. 2009; 11: 1811-1815.
- Sunkar S, Nachiyar CV. Microbial Synthesis and characterization of silver nanoparticles using the endophytic bacterium Bacillus cereus: A novel source in the benign synthesis. Global J. Med. Res. 2012; 2: 953-959.
- Mahdieh M, Zolanvari A, Azimee A, Mahdieh M. Green biosynthesis of silver nanoparticles by Spirulina platensis. Scientia Iranica. 2012; 19: 926-929.
- Tsibakhashvili N, Kalabegishvili T, Gabunia V, Gintury E, Kuchava N, Bagdavadze N, et al. Synthesis of Silver Nanoparticles Using Bacteria. Nano Studies. 2012; 2: 179-182.
- Saifuddin N, Wong CW, Yasumira AN. Rapid biosynthesis of silver nanoparticles using culture supernatant of bacteria with microwave irradiation. E-Journal Chem. 2009; 6: 61-70.
- Ahmad A, Mukherjee P, Senapati S, Mandal D, Khan MI, Kumar R, et al. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Collidal Surf. B. 2003; 28: 313-318.
- Ingle A, Rai M, Gade A, Bawaskar M. Fusarium solani: a novel biological agent for the extracellular synthesis of silver nanoparticles. J. Nanopart. Res. 2009; 11: 2079-2085.
- Birla SS, Tiwari VV, Gade AK, Ingle AP, Yadav AP, Rai MK. Fabrication of silver nanoparticles by Phoma glomerata and its combined effect against Escherchia coli, Pseudomonas aeruginosa and Staphylococcus aureus. Lett. Appl. Microbiol. 2009; 48: 173-179.
- Gajbhiye M, Kesharwani J, Ingle A, Gade A, Rai M. Fungus-mediated synthesis of silver nanoparticles and their activity against pathogenic fungi in combination with fluconazole. Nanomed. 2009; 5: 382-386.
- Bawaskar M, Gaikwad S, Ingle A, Rathod D, Gade A, Duran N, et al. A new report on mycosynthesis of silver nanoparticles by Fusarium culmorum', Current Nanoscience. 2010; 6: 376-380.
- Castro-Longoria E, Vilchis-Nestor AR., Avalos-Borja M. Biosynthesis of silver, gold and bimetallic nanoparticles using the filamentous fungus Neurospora crassa', Colloids and Surf. B. 2011; 83: 42-48.
- Vahabi K, Mansoori G, Karimi S. Biosynthesis of Silver Nanoparticles by Fungus Trichoderma reesei. Nanotechnol Insci. J. 2011; 1: 65-79.
- Gaikwad SC, Birla SS, Ingle AP, Gade AK, Marcato P, Rai MK, et al. Screening of different Fusarium species to select potential species for the synthesis of silver nanoparticles. J. Braz. Chem. Soc. 2013. Online published.
- Raut RW, Lakkakula JR, Kolekar NS, Mendhulkar VD, Kashid SB. Phytosynthesis of silver nanoparticle using Gliricidia sepium (Jacq.). Curr. Nanosci. 2009; 5: 117-122.
- Mude N, Ingle A, Gade A, Rai M. Synthesis of silver nanoparticles using callus extract of Carica papaya-A first report', J. Plant Biochem & Biotechnol. 2009; 18: 83-86.
- Saxena A, Tripathy RM, Singh RP. Biological synthesis of silver nanoparticles by using Onion (Allium cepa) extract and their antibacterial activity. Digest J. Nanomater & Biostr. 2010; 5: 427-432.
- Roy N, Barik A. Green Synthesis of Silver Nanoparticles from the Unexploited Weed Resources. Int. J. Nanotechnol & Appl. 2010; 4: 95-101.
- Bonde SR. A biogenic approach for green synthesis of silver nanoparticles using extract of Foeniculum vulgare and its activity against Staphylococcus aureus and Escherichia coli. Nusantara Biosci. 2011; 3: 59-63.
- Mallikarjun K, Narsimha G, Dillip GR, Praveen B, Shreedhar B, Lakshmi S, et al. Green synthesis of silver nanoparticles using Ocimum leaf extract and their characterization. Digest J Nanomater. Biostruct. 2011; 6: 181-186.
- Udayasoorian C, Kumar KV, Jayabalakrishnan RM. Extracellular Synthesis of Silver Nanoparticles Using Leaf Extract of Cassia Auriculata. Digest J. Nanomater and Biostructures. 2011; 6: 279 - 283.
- Jegan A, Ramasubbu A, Kumar VS, Balamurgan M, Saravanan S. Environmental Benign Synthesis And Characterization of Silver Nanoparticles Using Phyllostachys Sp. Leaves Extract. Digest J. Nanomater. & Biostructures. 2011; 6: 325-330.
- Ramgopal MC, Saisushma IH, Attitalla A, Alhasin AM. A facile green synthesis of silver nanoparticles using soap nuts. Res. J. Microbiol. 2011; 6: 432-438.
- Bonde SR, Rathod DP, Ingle AP, Ade RB, Gade AK, Rai MK. Murraya koenigii Mediated Synthesis of Silver Nanoparticles and Its Activity against Three Human Pathogenic Bacteria. Nanosci. Methods. 2012; 1: 25-36.
- Gupta A, Bonde SR, Gaikwad SC, Ingle AP, Gade AK, Rai MK. Lawsonia inermis-mediated Synthesis of Silver Nanoparticles: Activity against human pathogenic Fungi and bacteria with special reference to formulation of a antimicrobial nanogel. IET Nanobiotechnol. 2013.
- Bacelar-Nicolau P, Johnson DB. Leaching of pyrite by acidophilic heterotrophic iron-oxidizing bacteria in pure and mixed cultures. Appl Environ Microbiol. 1999; 65: 585-590.
- Dopson M, Lindstrom EB. Potential role of thiobacillus caldus in arsenopyrite bioleaching Appl Environ Microbiol. 1999; 65: 36-40.
- Parameswari E, Lakshmanan A, Thilagavathi T. Biosorption and metal tolerance potential of filamentous fungi isolated from metal polluted ecosystem. Electr. J. Environ. Agric. food chem. 2010; 9: 664-671.
- Reichenbach H, Höfle G. Myxobacteria as producers of secondary metabolites. In: Drug Discovery from Nature. Grabley S, Thiericke R, editors. Springer, Berlin; New York. 1999; 149 - 179.
- Reichenbach H, Dworkin M. The order Myxobacterales, In: Prokaryotes. Starr KP, Stolp H, Trüper HG, Belows A, Schleje HG, editors. Springer-Verlag, Berlin. 1981; 328-355.
- Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam.nov. Int. J. Syst. Bacteriol. 1996; 46: 1088-1092
- Shimkets LJ, Dworkin M, Reichenbach H. The myxobacteria. In: The Prokaryotes. Third Edition, Dworkin M, Falkow S, Rosenberg E, Schileifer KH, Stackebrandt E, editors. Springer Science+Business Media, LLC. 2006; 7: 31-115.
- Sondi I, Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J Colloid Interface Sci. 2004; 275: 177-182.
- Mock JJ, Barbic M, Smith DR, Schultz DA, Schultz S. Localized surface plasmon resonance effects by naturally occurring Chinese yam particles. J. Chem. Phy. 2002; 116: 6755.
- Raheman F, Deshmukh S, Ingle A, Gade A, Rai M. Silver nanoparticles: Novel antimicrobial agent synthesized from aendophytic fungus Pestalotia sp. isolated from leaves of Syzygium cumini (L.). Nano Biomed. And Eng. 2011; 3: 174-178.
- You C, Han C, Wang X, Zheng Y, Li Q, Hu X, et al. The progress of silver nanoparticles in the antibacterial mechanism, clinical application and cytotoxicity. Mol Biol Rep. 2012; 39: 9193-9201.
- Dar M, Ingle A, Rai M. Enhanced antimicrobial activity of silver nanoparticles synthesized by Cryphonectria sp. evaluated singly and in combination with antibiotics. Nanomedicine. 2013; 9: 105-110.
- Gurunathan S, Kalishwaralal K, Vaidyanathan R, Venkataraman D, Pandian SR, Muniyandi J, et al. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf B Biointerfaces. 2009; 74: 328-335.
- Huang J, Chen C, He N, Hong J, Lu Y, Qingbiao L, et al. Biosynthesis of silver and gold nanoparticles by novel sundried Cinnamomum camphora leaf. Nanotechnol. 2007; 18: 105104.1-105104.11.
- Sastry M, Ahmad A, Khan MI, Kumar R. Biosynthesis of metal nanoparticles using fungi and actinomycete. Curr sci. 2003; 85: 162-170.
- Lara HH, Ayala-Núñez, Turrent LCI, Padilla CR. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J. Microbiolol. Biotechnol. 2010; 26: 615- 621.
- Sintubin L, De Gusseme B, Van der Meeren P, Pycke BFG, Verstraete W, Boon N. The antibacterial activity of biogenic silver and its mode of action. Appl. Microbiol. Biotechnol. 2011; 91: 153-162.