Research Article
Austin J Biotechnol Bioeng. 2014;1(2): 6.
Genetic Fingerprinting of Antimicrobial Fluorescent Pseudomonads Associated with Banana Rhizosphere
Singh BP1*
1Department of Biotechnology, Mizoram University, India
*Corresponding author: Singh BP, Department of Biotechnology, Mizoram University, Aizawl, Mizoram-796004, India.
Received: July 02, 2014; Accepted: July 28, 2014; Published: July 30, 2014
Abstract
In recent years, fluorescent pseudomonads are well recognized for their plant growth promoting and bio-control activities. This study was design to study the distribution of potential antimicrobial fluorescent pseudomonads associated with banana rhizosphere by using ERIC and BOX-PCR fingerprinting. For this purpose, 32 fluorescent pseudomonads were isolated from banana rhizosphere collected from three different major grown banana locations and screened against three fungal phytopathogens. Promising isolates were selected for genetic fingerprinting by using ERIC and BOX-PCR to establish clonal relationship among the antifungal fluorescent pseudomonads. ERIC-PCR fingerprinting was found to be a potential tool to differentiate among the fluorescent and non fluorescent bacterial isolates as compare to BOX-PCR fingerprinting. A subset of 25 antagonistic fluorescent pseudomonads were characterized by ERIC and BOX-PCR and showed a high degree of DNA heterogeneity. Four groups were identified based on ERIC-PCR and three of them showed homogeneity containing fluorescent pseudomonas with a group of non fluorescent bacillus sp (PF 7). A unique band of 450 bp was generated by ERIC-PCR profiling among all the fluorescent isolates, which can be used as a diagnostic tool, whereas non fluorescent bacteria will have an adjacent band of 430 bp, which was absent in all identified fluorescent pseudomonas isolates. Further, 16S rDNA based identification of randomly selected isolates proved that ERIC-PCR fingerprinting is better DNA based tool to differentiate fluorescent pseudomonas with other bacterial isolates. We conclude and recommend ERIC-PCR banding pattern could be potential tool to differentiate among fluorescent and non fluorescent group of bacteria.
Keywords: Fluorescent pseudomonads; Genetic fingerprinting; Antibacterial; ERIC-PCR; BOX-PCR; Repetitive sequences
Abbreviations
PDA: Potato Dextrose Agar; PCR: Polymerase Chain Reaction; ERIC-PCR: Entrobacterial Repetitive Intergenic Consensus Polymerase Chain Reaction; EDTA: Ethylene Diamine Tetra Acetic acid; SDS: Sodium Dodecyl Sulfate; TAE: Tris Acetate EDTA; UPGMA: Unweighted Pair Group Method with Arithmetric average; 16S rDNA: 16S ribosomal DNA; NCBI: National Centre for Biotechnology Information; BLAST: Basic Local Alignment Search Tool
Introduction
The fluorescent pseudomonads, exist as one of the major group within the rRNA homology group I of genus pseudomonas, are characterized by the production of fluorescent pigment, produced on media containing low iron content and are responsible for the biodegradation of diverse range of compounds [1-3]. The group includes species for the production of bioactive compounds like, bio control agents of animal and plant diseases, promote plant growth, and control soil born diseases and production of plethora of biotechnologically important compounds [4-6]. Fluorescent pseudomonads were widely distributed which suggests their range of adaptability with the nature. This group of bacteria is preferably matched as soil inoculants due to aggressive colonization with the plant roots. This feature alone is suggested as a disease control mechanism by preventing the invasion of soil pathogens onto the root surface [7-11]. To apply these organisms in practice, it is verymuch important to understand the molecular diversity and metabolic versatility associated with different rhizosphere [12-14].
Banana is considered as one of the important food item due to its high nutritional and commercial values throughout the tropics of the world. It is the one of the major tropical fruit crop in 120 countries with annual production of 102 million tonnes [15]. Northeast India is considered as a rich source for the diversity of banana varieties which are believed to be hybrids of Indian subcontinents and Southeast Asia [16]. Wild and cultivated bananas are abundantly available in Mizoram, one of the states in Northeast India [17]. Henceforth, sustaining and enhancing the growth and productivity of banana have become a major focus of research. The growth and yield of banana in field, is greatly influenced by wide range of fungal and bacterial diseases. Control of diseases by the use of various chemicals and use of resistance cultivars are the important methods to improve the yield of crop plants. However, it is not possible to have resistant cultivar for every disease. Moreover, the availability of chemical fungicides is costly and has adverse effects on human health and is found to be lethal for the rhizospheric microorganisms. It has been recognized that the rhizospheric microorganisms are natural antagonistic towards important phytopathogens and offers us as an alternative tool against chemical fertilizers.
Genetic typing techniques based on conserve repetitive regions have been shown to be more accurate and discriminatory than morphological and phenotypic methods for typing pseudomonas isolates [18-20]. Rep-PCR fingerprinting utilizes primers from highly conserved repetitive sequences of the bacterial genome. Two of such type of elements is entrobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) and BOX-PCR more common to gram negative enteric bacteria [21]. The fingerprinting generated by using ERIC-PCR shows characteristic pattern and could be used to differentiate bacterial genomes [22].
Mizoram has been known for its diversified nature in the field of plant and animal biodiversities and can offer a wide range of research objectives in near future. Very limited research had been done so far in the local microbial biodiversity [23-25]. Majority of the population is living in rural areas and depends on agriculture. Lack of proper methods of agricultural farming and extensive use of chemical fertilizers are rapidly depleting the fertility of soils in cultivated lands. The use of chemical fertilizers and conventional agricultural practices such as jhumming could not sustain the fertility of the soil for long terms [26]. As a result, farmers are not able to get the worth of their hard works with fertility of soil rapidly decreasing and the yield of crop plants lowering day by day. Henceforth, the use of native microorganism to fight against soil born diseases will be a best alternative tool to increase crop production. This study was design to search for a potential antifungal fluorescent pseudomonas to develop a crop specific bio-fertilizer and to understand the genetic relationship among them by using repetitive sequence markers.
Materials and Methods
Isolation of fluorescent pseudomonads
Fluorescent pseudomonads were isolated from the rhizosphere of selected banana cultivated places in Aizawl around Mizoram, Northeast India. Three places were selected based on the production of banana in Mizoram and samples were collected randomly and pooled together before processing. Roots were collected with adhering soil and homogenized in washing buffer (0.1 M potassium phosphate, pH 7) solution followed by serial dilution and plating on King's B agar medium. After spreading, plates were incubated at 30°C for 24 hours, and colonies those fluoresced under UV light (λ= 356 nm) were selected and were further purified on same media by repetitive streaking.
Antimicrobial screening of fluorescent pseudomonads
Fluorescent pseudomonads were isolated from the rhizosphere of banana cultivated laces in Aizawl around Mizoram, Northeast India (23.44N 92.57E). All the isolates were screened for antifungal activities against three fungal phytopathogens viz. Fusarium oxysporum (CABI- 293942), Fusarium udum (MTCC-2755) and Fusarium graminearum (MTCC-1893) by dual plate assay [27]. All the pathogens were grown on PDA media and agar plug (4 mm diameter) was taken by using sterilized cork borer and placed on PDA agar plates. At the same time, bacterial cultures were streaked about 3 cm away from the agar plug towards the edge of the plates. A plate alone with fungal plug was used as a control and the plates were incubated at 28°C for 5 days. Inhibition diameter was recorded in mm against each pathogen.
Genomic DNA extraction and quantification
A single purified bacterial colony of the potential isolates showing significant antifungal activity at least against two phytopathogens was inoculated in 10 ml nutrient broth (Hi-Media, Mumbai, India) and incubated in orbital shaker for 24 hours at 30°C. Cells were precipitated by centrifugation and re-suspended in 2ml of 25% sucrose, 50mM EDTA (pH 8.0). Saturated lysozyme solution (150 μl) and 10 μl of 20 mg ml-1 RNAse A were added and incubated at 37°Cfor 3 hours. Lysis was done by using 150 μl of 20% SDS, mixed gently and incubated at 37°C for 10 minutes and suspension was incubated in water bath at 65°C for 2 hours. Other macromolecules like proteins and carbohydrates were removed by extraction with chloroform/isoamyl alcohol (24:1). The resulting suspension was mixed gently andcentrifuges at 10,000 rpm for 10 minutes and upper clear layer was transferred in new autoclaved tubes. DNA was precipitated by adding absolute alcohol and DNA was spooled out and dissolved in 200 μl of TE buffer (10mM Tris-Cl, pH-8.0; 1mM EDTA). Quantification of DNA was done by using UV-VIS spectrophotometer by taking the ratio at OD260/OD280.
ERIC and BOX-PCR fingerprinting
Universal primers based on ERIC sequences were used to generate fingerprinting were ERIC1 (5'-ATGTAAGCTCCTGGGGATTCAC-3') and ERIC2 (5'-AAGTAAGTGACTGGGGTGAGCG-3') while those used for BOX-PCR was BOXA1R (5'-CTACGGCAAGGCGACGCTGACG-3') [28]. The reaction mixture for ERIC-PCR after standardization consisted of 1 μl of DNA template, 0.6 μl dNTP mix (15mM), 100 pmol of primer ERIC1R and ERIC 2, 1 μl of MgCl2 (50mM) and unit of Taq DNA polymerase (Invitrogen- life technologies, USA),final volume was makeup to 25 μl by adding remaining sterilized water. Negative control reaction without template DNA was used for each amplified set. Amplification was conducted in Veriti Applied Bio-system thermal cycler with an initial denaturation step at 95°C for 7 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 51°C for 1 min, extension at 65°C for 8 min., followed by final extension at 65°C for 16 min. The reaction mixture for BOX-PCR is similar with ERIC-PCR except the annealing temperature for BOXPCR was 500C for 1 min. The products were analyzed by running on 1.5% agarose gel in 1X TAE buffer, 1kb DNA ladder (Merk pvt. Ltd) was used as size standard. The gel was stained by ethidium bromide (0.5 μg/ml), and visualized under a gel documentation system (Bio- Rad Gel Doc XR+, USA).
PCR amplification of 16S rDNA and sequencing
Randomly ten isolates were selected for identification by amplification of 16S rDNA region of bacterial rRNA genes with universal E. coli primers, forward from position 7 to 26 (5'-AGAGTTTGATCCTGGCTCAG-3') [29] and reverse primer from position 1,513-1492 (5'-ACGGCTACCTTGTTACGACTT-3') [30]. Amplification was performed in 20μl reaction mixture consisting of 2μl of 10X PCR assay buffer (10X), 2.0μl of dNTPs (2.5mm), 0.6μl primers (10 pmol), 0.2μl of Taq polymerase (1U/μl), 1.5μl of template DNA (50ng/ μl), and remaining of Millipore distilled water. Amplification was conducted in Veriti Applied Bio-system thermal cycler with an initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min 20 sec, followed by final extensionat 72°C for 6 min. Gel visualization was done as stated earlier. Sequencing was done commercially for the amplified products. Bioinformatics tool BLAST was used to locate the closest relative and sequences were submitted to NCBI Gen Bank.
Data analysis
The banding pattern generated by using ERIC and BOX-PCR were scored as absent (0) or present (1) and the resulting bands were used to generate similarity matrix for the construction of phylogenetic tree by NTSYS-pc analysis package [31]. Jaccard's coefficient of similarityindex was used to calculate similarity distances [32]. Cluster analysis was done using unweighted pair-group method with arithmetric average (UPGMA).
Results and Discussion
Isolation and antifungal screening of fluorescent pseudomonads
In total 32 isolates with characteristic fluorescent pigment were isolated from rhizosphere of banana collected from three different locations. All the isolates were screened for their antifungal potential against three fungal phytopathogens. Among them, 25 isolates, were shown significant activity against at least two fungal pathogens were selected for genetic fingerprinting by using ERIC and BOX-PCR and designated as PF1- PF25 (Table 1). Sixteen isolates inhibited all three pathogens whereas remaining nine isolates were inhibiting two pathogens. The inhibition zone was ranging from 5 to 18 mm in diameter (Table1). Fluorescent pseudomonads are one of the common culturable soil bacteria known to promote plant growth either by enhancing plant growth or suppressing the soil-borne pathogens. Their predominance in the rhizosphere of various plants is being reported, but genetic fingerprinting of potential fluorescent pseudomonads is in its initial phase [33-35]. This group is of interest due to their abundance in soil and their role in controlling different fungal pathogens [36]. A significant number of isolates (25 out of 32) exhibited antifungal activity against selected phytopathogenic fungi. Similar findings were reported by Neelamegam, 2012 [15] in the fluorescent pseudomonads isolated from sugarcane rhizosphere. Plant growth promoting traits of fluorescent pseudomonads was well studied under normal and saline conditions [6].
Isolate No.
NCBI Accession Number
Biochemical Characterization
Antifungal Screening (mm*)
Grams Reaction
Shape
Motility
Spores
FG
FO
FU
PF1
KM112023
-
Rod shaped
Motile
-
++
++
+++
PF2
NS
-
Rod shaped
Motile
-
+
++
-
PF3
KM112030
-
Rod shaped
Motile
-
++
+++
++
PF4
NS
-
Rod shaped
Motile
-
+
++
++
PF5
NS
-
Rod shaped
Motile
-
+
++
-
PF6
KM112031
-
Rod shaped
Motile
-
++
+++
++
PF7
KM112032
-
Rod shaped
Motile
-
+
-
++
PF8
NS
-
Rod shaped
Motile
-
++
++
++
PF9
KM112024
-
Rod shaped
Motile
-
+
++
++
PF10
NS
-
Rod shaped
Motile
-
+
-
++
PF11
KM112025
-
Rod shaped
Motile
-
++
++
+++
PF12
NS
-
Rod shaped
Motile
-
-
++
+
PF13
NS
-
Rod shaped
Motile
-
+
++
++
PF14
NS
-
Rod shaped
Motile
-
+
++
++
PF15
KM112026
-
Rod shaped
Motile
-
+
++
++
PF16
NS
-
Rod shaped
Motile
-
+
-
++
PF17
KM112027
-
Rod shaped
Motile
-
+++
+
++
PF18
NS
-
Rod shaped
Motile
-
++
++
-
PF19
KM112028
-
Rod shaped
Motile
-
++
+
++
PF20
NS
-
Rod shaped
Motile
-
+
-
++
PF21
KM112029
-
Rod shaped
Motile
-
++
++
++
PF22
NS
-
Rod shaped
Motile
-
+
++
++
PF23
NS
-
Rod shaped
Motile
-
+++
+++
++
PF24
NS
-
Rod shaped
Motile
-
++
++
-
PF25
NS
-
Rod shaped
Motile
-
++
++
++
Table 1: Biochemical and antifungal screening of fluorescent pseudomonads of banana rhizosphere.
Biochemical characterization of fluorescent pseudomonads
Phenotypic characterization of the banana rhizosphere isolates shows that all are gram-negative, non spore forming, rod shaped motile organisms. All the selected antifungal isolates were exhibiting catalase, oxidase and arginine hydrolysis activities (Table 1). Based on these characteristics, isolates could be identified as Fluorescent pseudomonas sp. Ten isolates based on ERIC-PCR pattern were randomly selected for identification by using 16S rDNA sequencing [37,35]. Biochemical characterization of banana rhizosphere isolates indicates that most of the isolates expected to be pseudomonas, but biochemical and phenotypic methods are not mare enough to distinguish and identified closely related isolates till species level [38,18]. This study is also shows the similar findings that biochemical and phenotypic methods could identify fluorescent pseudomonads till genus level, which is proved by 16S rDNA sequencing [39,40].
ERIC and BOX- PCR fingerprinting of fluorescent pseudomonads
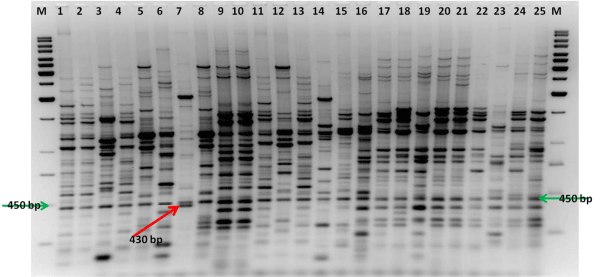
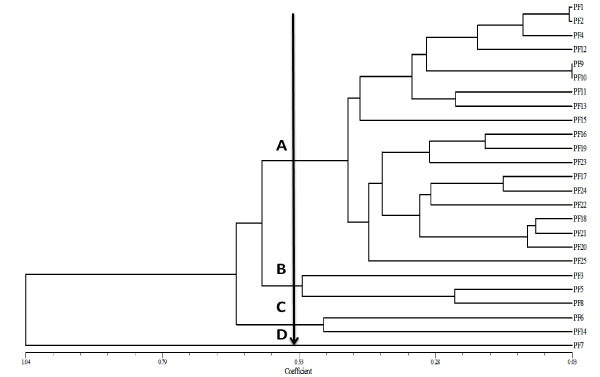
Whole genomic DNA, ERIC-PCR amplification of the antifungal fluorescent pseudomonad isolates yielded unique genomic fingerprinting pattern consisting of 10-31 products ranging in size from 150 bp to 3000 bp (Figure 1), which shows this technique was useful to differentiate the isolates. Dendrogram analysis was divided the isolates into four clusters (A-D) (Figure 2). Cluster A was the major cluster containing 19 isolates, out of which 7 were randomly selected for identification, among which two were identified as P. braccicacearum (PF 17:KM112027 and PF 21: KM112029) and one each as P. aeruginosa (PF 1: KM112023), P. Syringae (PF 9: KM112024), P. koreensis (PF 11: KM112025), P. Putida (PF 15: KM112026), Pseudomonas sp.(PF 19: KM112028) Cluster B was the second large cluster consists of 3 isolates, which were belongs to Pseudomonas sp. Cluster C and D contains 1 isolates each which were belongs to P. mendocina (PF 6: KM112031) and Bacillus sp. (PF 7: KM112032) respectively. Interestingly, isolate no PF 7, which was not having any similarity with other isolates was identified as Bacillus sp. We found that ERIC-PCR fingerprinting could make very clear-cut differentiation among the fluorescent pseudomonas and other genus as all the pseudomonas isolates were fall in other clusters and Bacilluswas placed in a separate cluster.
Figure 1: ERIC-PCR fingerprinting pattern of fluorescent pseudomonad isolates isolated from banana rhizosphere (Lane 1 to 25 are represented as isolate no PF 1 to PF 25); Lane M is a standard 1 kb DNA ladder; Highlighted unique bands of 450 bp and 430 bp.
Figure 2: ERIC-PCR generated Dendrogram showing relationship among fluorescent pseudomonad isolates. Cluster analysis was performed by UPGMA with a matrix calculated with Jaccard's coefficient.
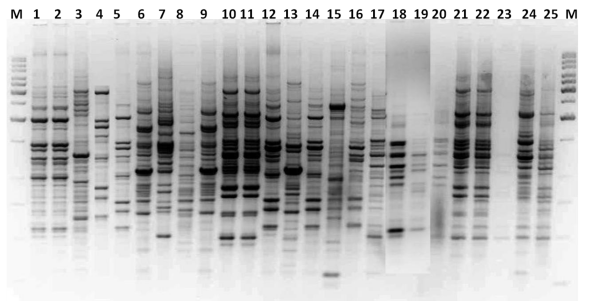
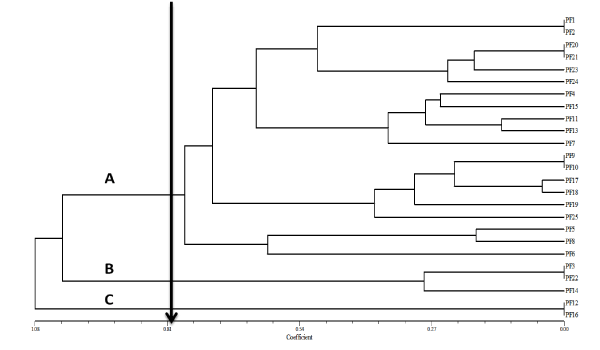
BOX-PCR fingerprinting pattern of the isolates differentiated the isolates belongs to banana rhizosphere, genomic fingerprinting pattern consisting of 06-25 products ranging in size from 250 bp to 2000 bp. However, it was not as discriminatory as ERIC-PCR fingerprinting (Figure 3). Dendrogram analysis divides the isolates into 3 clusters (A-C) (Figure 4). Cluster A was the largest cluster consists of 20 isolates and cluster B and C consists of 3 and 2 isolates respectively. In the box proofing the fluorescent pseudomonas isolates were mixed in different clusters and non fluorescent bacillus sp. (PF 7) also mixed with other fluorescent isolates (PF 11 & PF 15). We concluded that the BOX-PCR fingerprinting could not able to differentiate between fluorescent and non fluorescent pseudomonads.
Figure 3: BOX-PCR fingerprinting pattern of fluorescent pseudomonad isolates isolated from banana rhizosphere (Lane 1 to 25 is represented as isolate no PF 1 to PF 25); Lane M is a standard 1 kb DNA ladder.
Figure 4: BOX-PCR generated Dendrogram showing relationship among fluorescent pseudomonad isolates. Cluster analysis was performed by UPGMA with a matrix calculated with Jaccard's coefficient.
Repetitive sequence analysis like rep, ERIC and BOX were useful to understand the diversity among the antifungal fluorescent pseudomonads associated with banana rhizosphere. Although in this study BOX sequences have their own level of genetic resolution to distinguish the organisms, they were not sufficient to differentiation among the fluorescent and non fluorescent pseudomonads. A positive correlation was observed among the phenotypic and ERICPCR fingerprinting, this is in contradiction with other findings [18,41]. ERIC-PCR fingerprinting pattern clearly differentiate among the fluorescent pseudomonads falls in one major cluster and bacillus sp. being non fluorescent was clearly located in a separate cluster, which was not true with the case of BOX-PCR, which was in agreement with the findings of Bruijn [22]. On the whole, the ERIC-PCR fingerprinting could be a potential and reliable tool for the detection and differentiating among fluorescent pseudomonads.A unique band of 450 bp generated by ERIC-PCR fingerprinting could also used as a detection method for the primary detection of fluorescent pseudomonads, though it needs to be verify by some other molecular techniques whereas non fluorescent bacteria generates an extra band of 430 bp, which was not present in any of the fluorescent pseudomonas (Figure 3).
Conclusion
In this study we concluded that fluorescent pseudomonas isolates obtained from banana rhizosphere could be differentiated based on DNA typing tool ERIC-PCR fingerprinting as compare to BOX-PCR banding pattern. ERIC-PCR profiling clearly differentiates the non fluorescent bacterial isolate PF 7, identified as bacillus sp. which falls under a different cluster. We also found a unique band of about 450 bp in all the fluorescent pseudomonas whereas in non fluorescent pseudomonas an extra band of 430 bp was also present, which was not present in any of the fluorescent pseudomonas (PF 7). Henceforth, we also concluded that band of 450 bp in ERIC-PCR profilingcould be used as an identification tool to differentiate between fluorescent pseudomonas with other bacterial group. Further, these findings describe the diversity, plant-growth promoting traits, and antagonistic potential of banana rhizosphere-associated fluorescent pseudomonas and exploitation of their plant beneficial traits for sustainable agriculture.
Acknowledgment
BPS is thankful to Indian Council of Agricultural Research for funding under National project on Application of Microorganisms in Agriculture and Allied Sectors (AMAAS) to National Bureau of Agriculturally Important Microorganisms (NBAIM), Mau, India. We are also thankful to Department of Biotechnology, Government of India, New Delhi for establishment of State-Biotech Hub and Bioinformatics facility at Department of Biotechnology, Mizoram University. The authors thank Mr. Hardayanto Sulisto Putro, Research Associate, University of Aberdeen, Scotland, UK for the critical reading and the language editing of the manuscript.
References
- Palleroni NJ. Gram-negative, aerobic rods and cocci: family: 1. Pseudomonadaceae. In: Bergeys Manual of Systematic Bacteriology. Krieg NR, Holt JG, Editors. 1st edn. Williams and Wilkins, Baltimore. 1984; 1: 141-219.
- Palleroni NJ. Introduction to the family pseudomonadaceae. In: The Prokaryotes. Balows HG, Truper M, Dworkin W, Harder A, Schliefer K, Editors. 2nd edn. Springer-Verlag, New York. 1992; 2: 3071-3085.
- Ferret C, Sterckeman T, Cornu JY, Gangloff S, Schalk IJ, Geoffroy VA. Siderophore-promoted dissolution of smectite by fluorescent pseudomonas. Environmental microbiology reports. 2014.
- Pérez-Brandán C, Huidobro J, Grümberg B, Scandiani MM, Luque AG, Meriles JM, Vargas-Gil S. Soybean fungal soil-borne diseases: a parameter for measuring the effect of agricultural intensification on soil health. See comment in PubMed Commons below Can J Microbiol. 2014; 60: 73-84.
- Sharma A, Wray V, Johri BN. Rhizosphere Pseudomonas sp. strains reduce occurrence of pre- and post-emergence damping-off in chile and tomato in Central Himalayan region. See comment in PubMed Commons below Arch Microbiol. 2007; 187: 321-335.
- Anitha G, Kumudini BS. Isolation and characterization of fluorescent pseudomonads and their effect on plant growth promotion. See comment in PubMed Commons below J Environ Biol. 2014; 35: 627-634.
- Mishra DS, Kumar A, Prajapati CR, Singh AK, Sharma SD. Identification of compatible bacterial and fungal isolate and their effectiveness against plant disease. See comment in PubMed Commons below J Environ Biol. 2013; 34: 183-189.
- Martinez-Viveros O, Jorquera MA, Crowley DE, Gajardo GM, Mora ML. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci Plant Nutr. 2010; 10: 293-319.
- Berg G, Roskot N, Steidle A, Eberl L, Zock A, Smalla K. Plant-dependent genotypic and phenotypic diversity of antagonistic rhizobacteria isolated from different verticillium host plants. Appl. Environ. Microbiol. 2002; 68: 3328-3338.
- Lemanceau P, Alabouvette C. Suppression of fusarium wilts by fluorescent pseudomonads: mechanisms and applications. Biocontrol. Sci. Technol. 1993; 3: 219-234.
- Ines M, Yousra T, Asma BR, Sana K, Abdennasser H. Multi-traits of non-pathogenic fluorescent pseudomonas and evaluation of their potential as biocontrol agents. American Journal of Environmental Science. 2014; 10: 199-209.
- Xu GW, Gross DC. Selection of fluorescent Pseudomonads antagonistic to Erwinia carotovora and suppressive of potato seed piece decay. Phytopathology.1986; 76: 414-422.
- Palleroni NJ. Pseudomonas classification. A new case history in the taxonomy of gram-negative bacteria. See comment in PubMed Commons below Antonie Van Leeuwenhoek. 1993; 64: 231-251.
- Rainey PB, Bailey MJ, Thompson IP. Phenotypic and genotypic diversity of fluorescent pseudomonads isolated from field-grown sugar beet. See comment in PubMed Commons below Microbiology. 1994; 140: 2315-2331.
- FAO. Food and Agriculture Organization of the United Nations (Production of Crops 2010 data).
- Molina AB, Kudagamage C. The international network for the improvement of banana and plantain (INIBAP): PGR activities in south Asia. In. South Asia Network on Plant Genetic Resources (SANPGR) meeting held on December 9-11 at Plant Genetic Resources Center (PGRC), Peradeniya, Sri Lanka. 2002: 1-7.
- Lalrinfela PC, Robert Thangjam. Genome characterization of banana genetic resources of Mizoram, India. Sci Vis. 2012; 12: 32-36.
- Rameshkumar N, Ayyadurai N, Kayalvizhi N, Gunasekaran P. Genotypic and phenotypic diversity of PGPR fluorescent pseudomonads isolated from the rhizosphere of sugarcane (Saccharum officinarum L.). See comment in PubMed Commons below J Microbiol Biotechnol. 2012; 22: 13-24.
- Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. See comment in PubMed Commons below J Clin Microbiol. 1996; 34: 1129-1135.
- Rademaker JLW, de Bruijn FJ. Characterization and classification of microbes by REP-PCR genomic fingerprinting and computer assisted pattern analysis. In: Caetano-Anolles, G., Gresshoff, P.M. (Eds.), DNA Markers: Protocols, Applications and Overviews. Wiley, New York. 1997: 151-172.
- Syrmis MW, O'Carroll MR, Sloots TP, Coulter C, Wainwright CE, Bell SC, et al. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J Med Microbiol. 2004; 53: 1089-1096.
- de Bruijn FJ. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 1992; 58: 2180-2187.
- Kanakala S, Singh BP. Plectosphaerella cucumeria occurrences as a new root rot pathogen and p-solubilizer in north-eastern India. Archives Phytopatho. Plant Protec. 2013; 46: 2016-2018.
- Zothansanga, Lalhmachhuani, Kumar NS, Gurusubramanian G. PCR pathotyping of native Bacillus thuringiensis from Mizoram, India. Sci Vis. 2011; 11: 171-176.
- Rebecca LH, Zothansang, Singh BP, Gurusubramanian G, Kumar NS. DNA finger printing of Bacillus thuringiensis based on repetitive DNA sequences using ERIC-PCR. Sci Vis. 2011; 11: 147-154.
- Tawnenga, Shankar U. Evaluating second year cropping on jhum fallows in Mizoram, northeast India-Phytomass dynamics and primary productivity. J. Biosci. 1996; 21: 563-575.
- Rios JL, Recio MC, Villar A. Screening methods for natural products with antimicrobial activity: a review of the literature. See comment in PubMed Commons below J Ethnopharmacol. 1988; 23: 127-149.
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. See comment in PubMed Commons below Nucleic Acids Res. 1991; 19: 6823-6831.
- Gamaleroa E, Fracchiaa L, Cavalettoa M, Garbayeb J, Frey-Klettb P, Varesec GC, et al. Characterization of functional traits of two fluorescent pseudomonads isolated from basidiomes of ectomycorrhizal fungi. Soil Biol. Biochem. 2003; 35: 55-65.
- Bossis E, Lemanceau P, Latour X, Gardan L. The taxonomy of pseudomonas fluorescens and Pseudomonas putida: Current status and need for revision. Agronomie. 2000; 20: 51-63.
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. See comment in PubMed Commons below Mol Biol Evol. 1987; 4: 406-425.
- Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vandoise Sci Nat. 1908; 44: 223-270.
- Gardener Mcspadden BB, Schroeder KL, Kalloger SE, Raaijmakers JM, Thomashow LS, Weller M. Genotypic and phenotypic diversity of phlD-containing pseudomonas strains isolated from the rhizosphere of Wheat. Applied and Environmental Microbiology. 2000; 65: 1939-1946.
- Ayyadurai N, Naik PR, Sakthivel N. Functional characterization of antagonistic fluorescent pseudomonads associated with rhizospheric soil of rice (Oryza sativa L.). See comment in PubMed Commons below J Microbiol Biotechnol. 2007; 17: 919-927.
- Laguerre G, Rigottier-Gois L, Lemanceau P. Fluorescent Pseudomonas species categorized by using polymerase chain reaction (PCR)/restriction fragment analysis of 16S rDNA. See comment in PubMed Commons below Mol Ecol. 1994; 3: 479-487.
- Hu HB, Xu YQ, Chen F, Zhang XH, Hur BK. Isolation and characterization of a new fluorescent Pseudomonas strain that produces both phenazine-1-carboxylic acid and pyoluteorin. J. Microbiol. Biotechnol. 2005; 15: 86-90.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. See comment in PubMed Commons below J Bacteriol. 1991; 173: 697-703.
- Widmer F, Seidler RJ, Gillevet PM, Watrud LS, Di Giovanni GD. A highly selective PCR protocol for detecting 16S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Applied Enviro Microbiology. 1998; 64: 2545-2553.
- Ross IL, Alami Y, Harvey PR, Achouak W, Ryder MH. Genetic diversity and biological control activity of novel species of closely related pseudomonads isolated from wheat field soils in South Australia. Appl. Environ. Microbiol. 2000; 66: 1609-1616.
- Naik PR, Sahoo N, Goswami D, Ayyadurai N, Sakthivel N. Genetic and functional diversity among fluorescent pseudomonads isolated from the rhizosphere of banana. See comment in PubMed Commons below Microb Ecol. 2008; 56: 492-504.
- Misko AL, Germida JJ. Taxonomic and functional diversity of pseudomonads isolated from the roots of field-grown canola. See comment in PubMed Commons below FEMS Microbiol Ecol. 2002; 42: 399-407.