Review Article
Austin J Biotechnol Bioeng. 2014;1(2): 3.
Utilization of Type IIS Endonucleases in Molecular Cloning
Jiayou Zhang*
Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, USA
*Corresponding author: Jiayou Zhang, Department of Microbiology, Immunology and Molecular Genetics, University of Kentucky, Medical Bldg, Room MN416, 800 Rose Street, Lexington, KY 40515, USA.
Received: July 02, 2014; Accepted: July 29, 2014; Published: Aug 01, 2014
Type II endonucleases are powerful tools for molecular cloning. These endonucleases digest at a defined position within or close to their recognition sites. The Type II endonucleases used the most are those that digest DNA within their recognition sites, such as EcoRI (GAATTC) or HindIII (AAGCTT). Some literature refers to them as orthodox Type II endonucleases [1]. They usually recognize 6 or 8 bases as symmetrical sequences. A symmetrical sequence reads the same forward and backward. These endonucleases contain two identical subunits (homodimers). One binds a three (or four) base sequence while the other one binds to the other strand of DNA and the same three (or four) base sequence at the opposite direction, so that the two units are situated face to face and upside down in relation to each other [2]. The recognition and cleavage domains of these endonucleases are not organized as two distinguish part in their 3-D structure [2]. Orthodox Type II endonucleases cleave within their recognized sequences, creating a sticky end or a blunt. (The enzymes that recognize four bases will be not discussed here, because of the high frequency of their recognition sites, making it difficult to find a unique one within a DNA sequence.) EcoRI, a well-known orthodox Type II endonuclease, recognizes 5'-GAATTC-3', and digests within the sequence as
G /A A T T C
C T T A A/ G
resulting in a 5' overhang 5'-AATT.
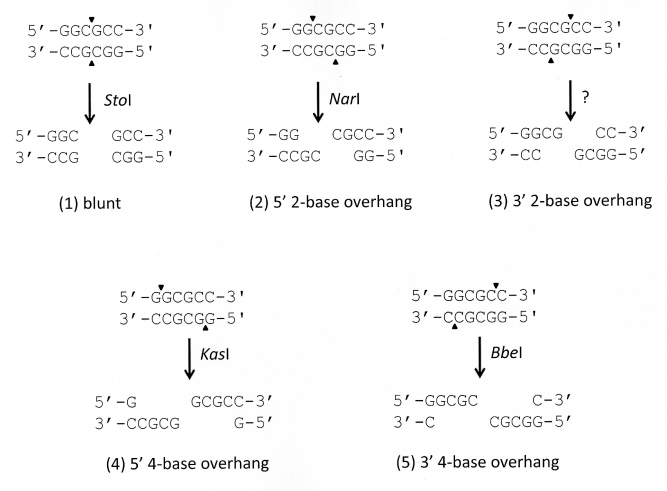
Digestion by orthodox Type II endonucleases that recognize a 6-base sequence makes it possible to create 5 different overhangs (Figure 1). So far, GAATTC is only recognized by EcoRI. However, the sequence GGCGCC could be recognized by 5 different orthodox Type II endonucleases: (1) Digestion with SfoI results in blunt end, and (2) NarI results in 5'-CG. An endonuclease that recognizes GGCGCC resulting in a (3) CG-3' overhang has not been discovered yet. Digestion with (4) KasI results in 5'-GCGC, and (5) BbeI results in GCGC-3'. These endonucleases that recognize the same sequence, but digest it differently are called "neoschizomers" [3].
Figure 1: The five possible overhangs created by digestion of 6-base symmetrical sequence with orthodox type II endonucleases.
In contrast, Type IIS endonucleases recognize asymmetrical sequences and digest outside of their recognition site [4]. Type IIS stands for a Type II endonuclease with an asymmetric target and cleavage sites [5].
The Type IIS endonuclease BsaI recognizes the 6-base sequence 5'-GGTCTC-3' and digests this sequence downstream of the site, creating 4-base sticky ends.
5'- G G T C T C N /N N N N -3'
3'- C C A G A G N N N N N/ -5'
Please note: This sequence can also be read as:
5'- /N N N N N G A G A C C -3'
3'- N N N N/ N C T C T G G -5'
The distance between the recognition and cleavage sequences is long for many commercially available Type IIS endonucleases which limits their utilization for manipulation of DNA. Type IIS endonucleases are monomeric (single proteins) [6]. The recognition domain and cleavage domain are separated from each other. For FokI, a Type IIS endonuclease, two domains even can be separated into two fragments by trypsin digestion [6]. The cleavage domain of FokI has been linked to different specific DNA binding proteins, resulting in "artificial" endonucleases that cut DNA close to their predetermined site [7].
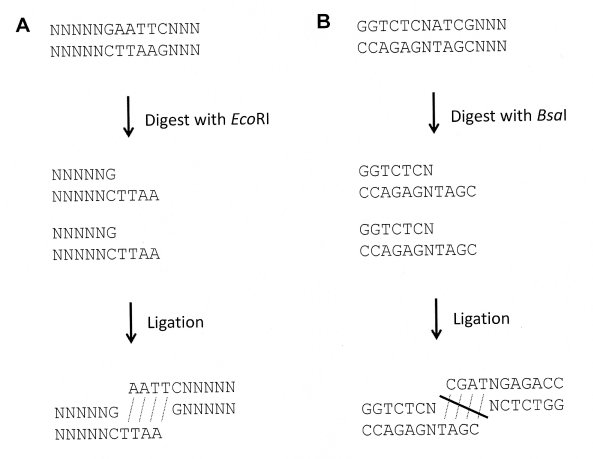
Type IIS endonucleases have not been used as popularly as orthodox Type II endonucleases. A significant reason for this is that the ends of EcoRI (a standard Type II endonuclease) are always compatible with ends of EcoRI. However, the ends created by BsaI, a Type IIS endonuclease, would be not compatible with ends of BsaI from a different sequence or even from an identical sequence, in most cases (Figure 2B). However, Type IIS endonucleases are very useful in molecular cloning, as more and more people prefer to generate insertion DNA fragments by PCR amplification.
Figure 2: Identical fragments digested with Type IIS endonucleases are not compatible. A. The overhangs resulting from digestion with orthodox endonucleases are symmetrical and compatible. B. Overhangs resulting from digestion with Type IIS endonucleases are usually not. These overhangs are not compatible and would not be ligated to each other
Since orthodox Type II recognition sequences of the four different kinds of overhangs are symmetrical, their overhangs exhibit symmetry too. Therefore, the ends digested by a specific standard Type II enzyme are compatible with its identical fragments (Figure 2A). In this case, an EcoRI-HindIII fragment A isolated using an agarose gel is designed to ligate a HindIII-EcoRI fragment B; only a small portion of ligations are between a single fragment A and single fragment B. Most ligations are between two identical fragments as or two fragment Bs, subsequent ligations result in a few very large fragments. As a result, most isolated fragments are wasted insteadforming a circle of DNA.
In contrast, the sticky ends created by Type IIS endonucleases are usually not symmetrical. The overhangs resulting from digestion with BsaI usually would not be not be able to ligate the identical 5'-ATCG overhang, because they are neither symmetrical nor compatible as shown in Figure 2B.
This BsaI end only can ligate with a fragment with the 5'-CGAT overhang created by BsaI or another Type IIS endonuclease such as BbsI, BsmBI, and so on.
For example,
G G T C T C N /A T C G N N N
C C A G A G N T A G C/ N N N (Fragment C)
digested with BsaI, results in:
5'-A T C G N N N
N N N (Fragment C digestion)
The overhang would not ligate with its identical overhang (Figure 2).
In the case of fragment D
5'-/C G A T N G A G A C C
3'- G C T A/ N C T C T G G (Fragment D)
digested with BsmBI, results in:
5'- C G A T N G A G A C C
N C T C T G G (Fragment D digestion)
Ligation between fragments C and D would not result in any head-to-head or tail-to-tail connection (if the other ends are not compatible). Therefore, ligations between two asymmetrical ends created by (the same or different) Type IIS endonucleases would not result in waste. Highly efficient ligation between two asymmetrical sticky-ends is very useful in molecular cloning. The general applications of Type IIS endonucleases are summarized below.
i. Ligation among multiple fragments
The efficiency of ligation between three fragments digested by orthodox endonucleases is low. Ligation between four is almost impossible to select for a correct connection, because most fragments ligate to the wrong one (Figure 2A). Since an asymmetrical overhang has to ligate to another, different asymmetrical overhang, the ligation efficiency between them can be almost 100% if the two fragments are equal in mole. Therefore, it would allow successful ligation among multiple fragments [8,9].
ii. Creation of a large library due to high efficiency of ligation
A fragment can be isolated by digestion, with one or two different Type IIS endonucleases creating two different asymmetrical overhangs. The two different overhangs can result from a single Type IIS endonuclease, because the Type IIS endonucleases cleavage sequences are outside their recognition sequence. These fragments are not ligated unless there are other fragments that contain two ends compatible with the first fragment. Since the second fragment would also not ligate itself, the product of ligation only can be between the two ones. Therefore, the ligation efficiency in this scenario would be greater than the one between fragments isolated by digestion with orthodox enzymes. It is possible to make a large plasmid library (>106) by using Type IIS endonucleases and electroporation technology (Zhang, unpublished data).
iii. Seamless connection of two sequences
Traditional cloning requires a connection between two fragment digestions by two enzymes with compatible ends. In most cases, if there is not a restriction site, it would be very easy to design a PCR primer containing a restriction site within the primer. However, if a project does not allow us to mutate the sequence, it would be not difficult to design a primer that contains a Type IIS restriction site. As mentioned above, the ATCG/CGTA created by BsaI can be replaced to any four nucleotide overhangs, making it possible to seamlessly connect any two sequences without the insertion or deletion of a single base [10].
iv. Creating an overhang compatible with ends digested by orthodox endonucleases
In case a cloning strategy is planned, a fragment is needed to insert between the EcoRI and HindIII sites within a plasmid vector; the fragment can be amplified by PCR with two primers. The first one contains an EcoRI site, while the second one contains a HindIII site. The PCR product then can be digested with the two endonucleases and the EcoRI-HindIII fragments can be used for insertion. However,attention must be paid to whether this fragment contains other EcoRI or HindIII sites, or the result would be in the digestion of this fragment into smaller pieces, making the product of ligation contain deletion (s). If there are more than one EcoRI or HindIII sites within this fragment, the first step can be checking whether there are EcoRI or HindIII compatible sites. The MfeI site (CAATTC) is compatible with the EcoRI site (GAATTC); both enzymes create the 5'- AATT overhang. No enzymes however, are compatible with HindIII (AAGCTT). If this fragment contains additional HindIII site (s), a 4-base 5' overhang of Type IIS endonucleases can be used to create the 5' overhang. There are at least three Type IIS endonucleases that recognize 6 bases (Table 1); in most cases, at least one from the three enzyme recognition sites would not be within this fragment. It is possible to design a primer that contains one of the three Type IIS sites. The digestion sites can be designed as 5'-AGCT (compatible with HindIII digestion) and the recognition sites can be designed outside the digested fragment. The fragment can be digested with EcoRI and the Type IIS endonuclease (instead of HindIII) and is used to insert between the EcoRI and HindIII sites of the plasmid. In addition, a choice can be made if the ligation site can be re-digested with HindIII by adding an A before the AGCT overhangs. If it is preferred that the ligation site not be digested with HindIII, the A can be replaced with T, C or G. Many Type IIS endonucleases are commercially available now. There are 5 possibilities resulting from digestion with an orthodox restriction enzymes described above (Figure 1). Commercially available Type IIS endonucleases can create the 4 possible overhangs resulting from any orthodox endonucleases except the 3' 4 base overhang. BstXI is not a Type IIS but Type IIP endonuclease. Digestion with BstXI results in a 3' 4 base overhang; meanwhile three bases have to be added to the PCR fragment, which probably can serve most purposes.
3' end 2 base overhang:
BsrDI
G C A A T G N N /
C G T T A C / N N
BtsI
G C A G T G N N /
C G T C A C / N N
5' end 2 base overhang:
FauI
C C C G C N N N N / N N
G G G C G N N N N N N /
5' end 3 base overhang:
BspQI
G C T C T T C N / N N N
C G A G A A G N N N N /
EarI
C T C T T C N / N N N
G A G A A G N N N N /
5' end 4 base overhang:
BsaI
G G T C T C N / N N N N
C C A G A G N N N N N /
BsmBI
C G T C T C N / N N N N
G C A G A G N N N N N /
BbsI
G A A G A C N N / N N N N
C T T C T G N N N N N N /
blunt
MlyI
G A G T C N N N N N /
C T C A G N N N N N /
3' end 4 base overhang:
BstXI*
C C A N / N N N N N T G G
G G T N N N N N / N A C C
Table 1: Most useful Type IIS endonucleases.
Several factors need to be considered to choose a Type IIS endonuclease during DNA cloning. First, it is important that the recognition sequences will not be very far away from the digestion sequence, so that the primer containing a Type IIS restriction site will not be very long. Second, the recognition sequence will not be very short, with at least 5 bases or more, so that their restriction sites are less frequent within a DNA fragment. And finally, different cases may call for choices between different types of overhangs.
The most useful Type IIS enzymes are shown in Table 1.
Acknowledgment
We thank Jennifer F. Rogers for helpful comments on this manuscript. This work was supported by the Markey Cancer Center, University of Kentucky.
References
- Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. See comment in PubMed Commons below Nucleic Acids Res. 2001; 29: 3705-3727.
- Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. See comment in PubMed Commons below Science. 1990; 249: 1307-1309.
- Chaudhuri K. Recombinant DNA Technology TERI Press. 2013.
- Szybalski W, Kim SC, Hasan N, Podhajska AJ. Class-IIS restriction enzymes--a review. See comment in PubMed Commons below Gene. 1991; 100: 13-26.
- Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. See comment in PubMed Commons below Nucleic Acids Res. 2003; 31: 1805-1812.
- Li L, Wu LP, Chandrasegaran S. Functional domains in Fok I restriction endonuclease. See comment in PubMed Commons below Proc Natl Acad Sci U S A. 1992; 89: 4275-4279.
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. See comment in PubMed Commons below Proc Natl Acad Sci U S A. 1996; 93: 1156-1160.
- Peng L, Liang X, Baek C-H, Clark J, Katzen F. One-Step Cloning. Cloning Made Easy Using Type IIS Restriction Endonucleases. Tutorials. 2013; 33: 17.
- Yount B, Denison MR, Weiss SR, Baric RS. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. See comment in PubMed Commons below J Virol. 2002; 76: 11065-11078.
- Padgett KA, Sorge JA. Creating seamless junctions independent of restriction sites in PCR cloning. See comment in PubMed Commons below Gene. 1996; 168: 31-35.