Research Article
Austin J Biotechnol Bioeng. 2014;1(3): 5.
Griseofulvin Producing Endophytic Nigrospora Oryzae from Indian Emblica Officinalis Gaertn: a New Report
Rathod DP, Dar MA, Gade AK and Rai MK*
Department of Biotechnology, Sant Gadge Baba Amravati University, India
*Corresponding author: Rai MK, Department of Biotechnology, Sant Gadge Baba Amravati University, Amravati¬-444602, Maharashtra, India.
Received: August 25, 2014; Accepted: September 19, 2014; Published: September 22, 2014
Abstract
Emblica officinalis (Amla) is widely used medicinal plant and is believed to increase defence against various types of diseases. E. officinalis harbours a range of endophytes responsible for synthesis of bioactive compounds. We screened endophytic fungi isolated from E. officinalis for their antimicrobial activity. During screening we found that a griseofulvin-producing endophytic fungus (Nigrospora oryzae) which is active against human pathogenic microbes. Nigrospora oryzae (DBT-150) was isolated and identified based on the morphology and ITS-rDNA sequence comparison. The presence of Griseofulvin was confirmed by standard spectra of Griseofulvin available in Mass Spectroscopy instrument. Griseofulvin is a non-toxic antifungal drug derived from several species of Penicillium. In the present study, these active compounds demonstrated activity against human pathogenic bacteria and fungi. This is a first report of Griseofulvin producing endophytic Nigrospora oryzae isolated from E. officinalis.
Keywords: Endophyte; Griseofulvin; Human pathogenic microbes; Nigrospora oryzae; Emblica officinalis
Introduction
Emblica officinalis is an important medicinal plant belonging to family Euphorbiaceae, also named as Amla, or Indian gooseberry. E. officinalis is native of India, but also grows in tropical and sub-tropical regions of Pakistan, Uzbekistan, Sri Lanka, South East Asia, China and Malaysia. Plant parts of E. officinalis is widely used in medicinal and effective against a range of diseases like cancer, heart disease, ulcer, diabetes, liver treatment, anaemia and various other diseases. The plant is also used as antioxidant, immune-modulator, antipyretic, analgesic, cyto-protective, antitussive and gastroprotective. It is also used in ophthalmic disorders, memory enhancing, lowering cholesterol level and neutralizing snake venom [1]. Medicinal plants harbor a variety of endophytes, which in turn protects their host from infections and grazing agents and also provide compliance to survive in unfavourable environmental conditions. Endophyte produces metabolites, which are being recognized as multipurpose compounds of antimicrobial agents [2].
Fungal endophytes produce metabolites, which are not only essential for the plant protection but are also important as medicines. The demand for new and highly effective bio control agents to control pathogens is huge, and the removal of synthetic compounds from the market is also demanding, because of their toxicity to the environment. Griseofulvin, an orally active antifungal drug is obtained from several species of Penicillium, Xylaria, Nigrospora, etc. It is an antifungal drug used for both animals and humans to treat various fungal diseases. Griseofulvin has been used as an antifungal antibiotic for the treatment of mycotic diseases of humans and veterinary animals [3]. Sommart et al. [4] reported three new hydronaphthalenone derivatives and one new dihydroramulosin derivative isolated from an endophytic fungus PSU-N24. Further they also studied the antifungal activity of griseofulvin against Microsporum gypseum SH-MU-4 with a minimum inhibitory concentration (MIC) of 2m g/ml. Park et al. [5] reported griseofulvin from an endophytic fungus Xylaria species. The authors also evaluated the efficacy of Griseofulvin against plant pathogenic fungi. Xia et al. [6] reported two new derivatives of an antifungal compound griseofulvin from the mangrove Kandelia candel (L.) endophytic fungus Nigrospora species collected from the South China Sea. They also studied the antitumor and antimicrobial activity of isolated compounds. The derivatives of griseofulvin are highly active and are known to treat various pathogenic and non-pathogenic disorders. Richardson et al. [7] reported potent antifungal agent griseofulvin extracted from endophytic Xylaria sp. was isolated from Pinus strobus and Vaccinium angustifolium.
In the present study, we isolated Nigrospora oryzae endophyte from leaves of E. Officinalis which secreates derivatives of Griseofulvin and checked its activity against human and plant pathogenic bacteria and fungi. This is the first report of endophytic N. oryzae isolated from E. officinalis.
Material and Methods
Experimental material
Healthy twigs and leaves of Emblica officinalis were collected from local forest of Amravati, Maharashtra, India. The endophytes were isolated according to modified method described by Strobel [8]. Leaves were cut into small segments (0.5-1 cm long), surfacesterilized with 4-5 % Sodium hypochlorite solution followed by 70% (v/v) ethanol, respectively, for 1 min, and then washed thrice with sterile distilled water. After surface sterilization, segments were dried and inoculated on potato dextrose agar, incubated at 25 ±2°C in the dark and observed daily for growth. The grown mycelia were purified on fresh potato dextrose agar and stored on PDA slants at 4°C for further study.
Identification of fungal endophyte using morphological and molecular markers
For the microscopic identification, the fungus was grown on the PDA and then identified on the basis of morphological and cultural characteristics. The cultures were used for DNA isolation followed by molecular identification. Isolation of DNA was performed by Chromous kit (Chromous Pvt Ltd Bangalore). The primers used for ITS amplification were ITS1 (5`- TCC GTA GGT GAA CCT GCG G-3`) and ITS4 (5`- TCC TCC GCT TAT TGA TAT GC-3`) [9]. Mastercycler personal (Thermal cycler, Eppendorf, North America) was employed in a thermal cycle for an initial denaturation at 94OC for 2 min followed by 34 cycles of 94OC for 30s, (ITS) (18S rDNA) and 72OC for 2 min, with a final extension at 72OC for 5 min. Polymerase chain reaction (PCR) products were sent to Chromous Biotech Pvt Ltd Bangalore India for sequence analysis.
Phylogenetic analysis
After sequencing, nucleotide sequences were compared with ITS sequence data strains available at the public databases Genbank (https://www.ncbi.nem.nih.gov) by using the BLASTN sequence match routines. The sequences were aligned using the CLUSTAL W program and phylogenetic and molecular evolutionary analyses were performed using MEGA6 [10]. The phylogenetic reformation was performed using the neighbour joining (NJ) algorithm, with bootstrap values calculated from 1000 replicate runs, using the software routines included in the MEGA software.
Extraction of crude extract from Nigrospora oryzae
The plugs of agar supporting mycelium growth were cut and transferred aseptically to a 500 mL Erlenmeyer flasks containing 200 mL potato dextrose broth with 2% dextrose and potato extract. The flasks were incubated at 280C on a rotary shaker (120 rpm) for 21 days for further growth. After mycelium growth 200 mL culture was filtered through cheese cloth. The filtrate was concentrated at 45-500C and extracted thrice by shaking with equal volume of Ethyl acetate. Finally, we obtained a gum like texture as a crude extract in Ethyl acetate solvent.
Antimicrobial activity of Nigrospora oryzae extracts
Antimicrobial activity of extract of N. oryzae was evaluated against two human pathogenic bacteria such as S. aureus (ATCC- 25923) and E. coli (ATCC-39403)], two human pathogenic fungi like Trichophyton mentagrophytes (NCIM-3100)] and Microsporum canis and one plant pathogenic fungus-Fusarium oxysporum. The antimicrobial assay was performed in triplicate. Bacteria were grown on preliminary nutrient agar medium (Hi-Media), human pathogenic fungi on Sabouraud Dextrose Agar (HiMedia) and plant pathogenic fungi were grown on PDA (Potato Dextrose Agar) (HiMedia). The antibacterial activities of extract were determined by disk diffusion method. To determine the combined effects, each standard antibiotic disk impregnated with 20 μL solution of extract was placed onto the nutrient agar plate inoculated with test pathogenic bacteria and then incubated at 37°C for 24 hours. A similar procedure was followed for the human pathogenic fungi. Sterile disks impregnated with 20 μL solution of extract were placed on Sabouraud Dextrose Agar medium plates inoculated with human pathogenic fungi and for F. oxysporum 20 μL of extract was places on PDA incubated at 27°C for seven days. Zones of inhibition were measured and compared with a standard antibiotic disk and griseofulvin solution.
Spectroscopic analysis
MS analyses were carried out using an electro-spray ionization-ion trap (ESI-IT) mass spectrometer. Mass analyses were conducted in positive mode with initial source parameters as (source voltage = 4.5 kV; sheath gas flow rate = 60 nitrogen, arbitrary units; capillary temperature = 160°C; capillary voltage = 10 V; tube lens = 5 V; octopole 1 voltage = -2.5V; octopole 2 voltage = -6V; interoctopol lens = -16V). All the spectra were obtained and analysed by LCQ Xcalibur software (Finnigan, Atlanta, GA, USA). Total current ion Mass spectra were obtained between a range of m/z 80 and m/z 1000. Charge state and isotopic distribution were analysed by a narrow-scan range mode (Zoomscan mode) and compared with isotopic profile calculation performed with Isopro software. Scanning was recorded for about 0.5 min corresponding to 15 scans. Spectra were developed in shape mode with a boxcar smoothing of 7 points. Concerning MS analysis, fungal extracts and pure griseofulvin solutions were prepared at different concentrations (100μg/ml and 10μg/ml respectively) in a mixture of methanol/water 95:5 (v/v) with 0.1% of trifluoro-acetic acid. These solutions were infused directly into the ESI probe with a 250-μl automated syringe (Hamilton, Reno, NV, USA) at a flow-rate of 3μl/min.
Statistical analysis
To analyze whether there is any significant difference in zone of inhibition between extract and combinations, we used 'one way ANOVA' for the relative variability within the separate classes of the experiment.
Results and Discussion
Identification of fungal endophyte (DBT-150)
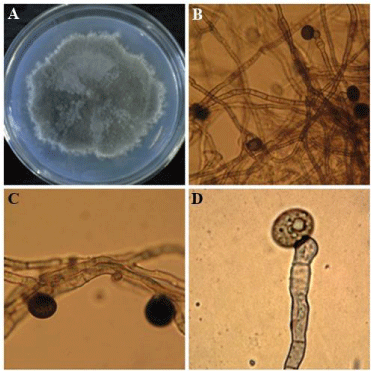
The fungal endophytes DBT-150 was identified as Nigrospora oryzae on the basis of morphology and molecular analysis (Figure 1). The genus Nigrospora is classified in family Trichosphaeriaceae of the class Ascomycota. Nigrospora species are reported as endophytes from different medicinal plants like Kandelia candel and Abies holophylla. The grey woolly colony grows fairly rapid. The diameter of colony was 5.5 cm after 7 days of incubation on PDA.
Figure 1: A: Morphology of Nigrospora oryzae on Potato Dextrose Agar, B: microscopic view of seperate mycelia and conidia, CD: enlargement of mycelia and conidia produced on swollen conidiophores.
The spores were produced singly on swollen urn-shaped conidiophores and egg-shaped to flattened-spherical, black (15μm), and often have an equatorial colourless line or germ slit. Usually it occurs as parasite on living grasses but also present on dead materials. It can be easily isolated from dead lawn grass in the autumn [11]. In the present study the Internal Transcribed Spacer (ITS) sequences demonstrated that the DBT-150 strain shows 100 % sequence similarity with Nigrospora oryzae of NCBI GenBank.
The endophyte isolated from E. officinalis was identified as N. oryzae both by morphological and molecular characterization based on rDNA gene sequences. Identification of Nigrospora sp., at the sub-genus level by morphological traits is difficult because of uneven shape and size of conidia all highly dependent on environmental conditions and composition of the media. Molecular techniques based on rDNA genes have been successfully applied to identify fungi particularly variable species of fungi [12]. Most species of Nigrospora are soil-borne and commonly distributed in nature as saprophytes and pathogens. Based on rDNA sequences the endophytic Nigrospora sp. shared a common clad with other isolates of N. oryzae already reported from various environments, confirming the identity of the endophytic fungal isolate as N. oryzae.
NCBI-BLAST and phylogenetic analysis
The BLASTN analysis of DBT-150 ITS1 sequence against nucleotide sequence collection showed the maximum identity with N. oryzae (HQ607943.1) and N. oryzae (HQ607925.1) with the similarity of 100 % and the least E-value of zero (Table 1). Based upon BLAST analysis showing the significant hits against the N. oryzae it is evident that the query sequence also belongs to N. oryzae.
Morphological Identification
Sequence based identification
Sequence with five best match
Identity (%)
Nigrospora sp.
Nigrospora oryzae
Nigrospora oryzae –FR872725.1
100
Nigrospora oryzae-KF192823.1
98
Nigrospora oryzae -HQ607943.1
98
Nigrospora oryzae- HQ607925.1
98
Nigrospora oryzae-JX129188.1
98
Table 1: Identification based on comparison of ITS sequences of endophytic Nigrospora oryzae using BLAST with sequences available in GenBank and their five best matches.
The phylogenetic analysis was carried out using ITS sequence region. Corresponding phylogenetic tree was generated with sequences from the different isolates obtained in this study together with those of other Nigrospora species from database. BLAST comparison of the sequences gave a similarity (>90%) within each set. For each locus, the closest sequences were those from the genus Nigrospora formed a close clad (Figure 2).
Figure 2: Phylogenetic relationship of the endophytic Nigrospora oryzae (DBT-150) isolated from Emblica officinalis to other fungi from GeneBank assume from sequences nuclear ribosomal ITS1-5.8S-ITS4.
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model [13]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 1.1286)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 11.5456% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 49 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 112 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [14].
Isolate sequences representing species from NCBI database and the sample sequences formed separate clades. The fungal endophytic isolate from Indian E. officinalis grouped in same clade of Nigrospora (Figure 2). Likeliness were most pronounced between the isolate sequences from endophytic fungi of E. officinalis (India) and those obtained from NCBI GeneBank, suggesting these isolates from India represents a new species in genus Nigrospora. This molecular and phylogenetic data is also supported by morphology of the isolates. In the present study we used universal ITS PCR primers for ITS based phylogenetic study, because it showed best results as compared to other primers fungi specific primers. As it is universal primer which easily amplify to any DNA of fungi. Due to that there is no need to use fungi specific primers for identification.
Antimicrobial activity of griseofulvin produced by Nigrospora oryzae
Antimicrobial activity of extract of endophytic N. oryzae was screened against human and plant pathogenic fungi like T. mentagrophytes, M. canis, F. oxysporum respectively and human pathogenic bacteria S. aureus (ATCC-6538), E. coli (ATCC-11775) by disc diffusion method. The MIC of extract was found to be 1.5 mg/ml as shown in Table 2. Griseofulvin extracted from N. oryzae DBT-150 inhibited the growth of both human and plant pathogenic fungi and bacteria. The growth of pathogenic fungi F. oxysporum, T. mentagrophytes, M. canis and bacteria S. aureus and E. coli were inhibited at less than 1.5, mg/ml.
Sr. No.
Organisms
Extract (mm)
Antibiotics* (mm)
Antibiotics* + Extract (mm)
Control
1
E. coli
11.66±0.57
21,66±0.57
22.66±0.57
0
2
S. aureus
11±1.0
20±1.0
21.66±1.52
0
3
Trichophyton mentagrophytes
21.33±2.51
11.33±1.15
22±2.0
0
4
Microsporum canis
19.66 ±0.57
14±2
21.66±1.52
0
5
Fusarium oxysporum
14 ±2.0
13.33±0.57
21.66±1.52
0
Table 2: Antibacterial and antifungal activity of Griseofulvin extracted from Nigrospora oryzae.
Characterization of griseofulvin compound by GC-MS
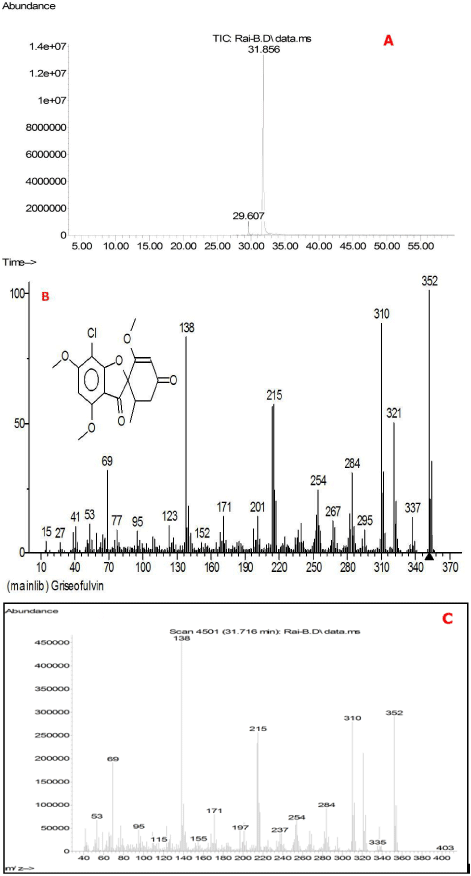
The extract of endophytic N. oryzae showed the presence of Griseofulvin by chromatographic techniques. Qualitative total ion chromatogram of the extract of the N. oryzae showed (Figure 3 A) the relative retention time (31.856 min). Further evidence for the identification of griseofulvin compound was determined by Mass Spectroscopy (MS) (Figure 3 B, C). The ionic composition analysis of the sample was analyzed by ESI-IT/MS and then compared with pure form. In mass spectra although the scan was very complex but a prominent monocharged peak was detected at m/z 353 to discriminate between the active fraction and others. The isotopic distribution revealed the presence of one chlorine atom.
Figure 3: A: Total ion chromatogram of Griseofulvin, B: mass spectra of the standard Griseofulvin and C: mass spectra of crude extract of N. oryzae extracted in Ethyl acetate shows similar molecular weight as standard griseofulvin compound.
The griseofulvin (Figure 3C) contains the same molecular weight (= 352 Da), contains a chlorine atom, depicting antifungal activity against human pathogenic microorganisms. Also, a comparison between obtained peak and pure griseofulvin has been performed. For this comparison a pure griseofulvin solution (10 μg/ml) was directly injected into the ESI-IT/MS, showed a major peak at m/z 353 corresponding to the protonated griseofulvin [M+H]+ with the same isotopic distribution compared to the observed unknown 353 peak in full scan of MS. The parameters for ESI were automatically optimized to obtain the best signal. 27 % collision energy was required for MS/MS analysis of griseofulvin. At m/z 215 a well differentiated chlorinated product ion appeared associated with intense ions at m/z 321 with chlorinated, relative abundance 48%, 310 (chlorinated, 85%), 284 (chlorinated, 30%) and 138 (non-chlorinated, 85%). In the same conditions, the unidentified m/z 353 ion gave the same product ions confirming that this peak could be attributed to griseofulvin.
There has been a pressing need to search for new antimicrobials from plants and microbes. Our study brings here new evidence of antimicrobial metabolite griseofulvin from N. oryzae isolated from E. officinalis. This antifungal agent griseofulvin extracted from N. oryzae would be the promising anti-fungal agent against human pathogens.
Pharmaceutical, industrial and agricultural bioactive metabolites such as antibiotics, antiviral, anticancer and antioxidant compounds are continuously reported and characterized from fungal endophytes. The reason why these bio products are bioactive compounds is not known, but it is understood that many of them act as chemical defence. As a result, biosynthesis of antifungal metabolites could be induced in response to other fungal competitors, thus providing a selective advantage for their development in natural environment. Griseofulvin is an antifungal compound isolated from different fungal species like terrestrial and marine species of Penicillium [15-16].
Mass spectroscopy is a powerful technique used to identify compounds in a complex extract with a high degree of conviction, which could not be obtained with classical methods such as TLC or HPLC-UV [17]. This study is a characteristic application of ESI/MS providing mass profiles of fungal extracts. After fractionation of the crude extract guided by an antifungal bioassay, ESI-IT/MS allowed the identification of griseofulvin as an active compound of N. oryzae.
Conclusion
An antifungal agent Griseofulvin isolated from endophytic fungus N. oryzae would be the promising antibacterial agent against human and plant pathogens. It can be concluded that endophytes act as an outstanding source for active bioactive compounds.
Acknowledgement
We are thankful to Ministry of Environment and Forest, New Delhi for providing financial support for project work. We also thank Dr. Marta Cristina Teixeira Duarte CPQBA-Centro Pluridisciplinar de Pesquisas Químicas, Biológicas e Agrícolas, Divisão de Microbiologia-Coordenadora, UNICAMP-Universidade Estadual de Campinas for help in characterization of griseofulvin.
References
- Khan KH. Roles of Emblica officinalis in Medicine - A Review. Bot Res Internat. 2009; 2: 218-228.
- Strobel GA. Rainforest endophytes and bioactive products. Crit Rev Biotechnol. 2002; 22: 315-333.
- ROTH FJ Jr. Griseofulvin. Ann N Y Acad Sci. 1960; 89: 247-253.
- Sommart U, Rukachaisirikul V, Sukpondma Y, Phongpaichit S, Sakayaroj J, Kirtikara K, et al. Hydronaphthalenones and a dihydroramulosin from the endophytic fungus PSU-N24. Chem Pharm Bull (Tokyo). 2008; 56: 1687-1690.
- Park JH, Choi GJ, Lee HB, Kim KM, Jung HS, Lee SW, et al. Griseofulvin from Xylaria sp. strain F0010, and endophytic fungus of Abiesholophylla and its antifungal activity against plant pathogenic fungi. J Microbiol Biotech. 2005; 15: 112-117.
- Xia X, Li Q, Li J, Shao C, Zhang J, Zhang Y. Two new derivatives of griseofulvin from the mangrove endophytic fungus Nigrospora sp. (strain No. 1403) from Kandelia candel (L.) Druce. Planta Med. 2011; 77: 1735-1738.
- Richardson SN, Walker AK, Nsiama TK, McFarlane J, Sumarah MW, Ibrahim A, et al. Griseofulvin-producing Xylaria endophytes of Pinus strobus and Vaccinium angustifolium: evidence for a conifer-understory species endophyte ecology. Fun Eco. 2014; 11: 107-113.
- Strobel G, Yang X, Sears J, Kramer R, Sidhu RS, Hess WM, et al. Taxol from Pestalotiopsis microspora, an endophytic fungus of Taxus wallachiana. Microbiology. 1996; 142: 435-440.
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a guide to methods and applications. Academic Press, New York, USA. 1990; 315-322.
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004; 5: 150-163.
- Barnett HL, Hunter BB. Illustrated Genera of Imperfect Fungi. 3rd edn. United State of America. Academic Press, London. 1972; 1-241.
- Cai L, Jeewon R, Hyde KD. Phylogenetic evaluation and taxonomic revision of Schizo- thecium based on ribosomal DNA and protein coding genes. Fun Div. 2005; 19: 1-17.
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993; 10: 512-526.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30: 2725-2729.
- Oxford AE, Raistrick H, Simonart P. Studies in the biochemistry of micro-organisms: Griseofulvin, C(17)H(17)O(6)Cl, a metabolic product of Penicillium griseo-fulvum Dierckx. Biochem J. 1939; 33: 240-248.
- Namikoshi M, Kobayashi H, Yoshimoto T, Meguro S, Akano K. Isolation and characterization of bioactive metabolites from marine-derived filamentous fungi collected from tropical and sub-tropical coral reefs. Chem Pharm Bull (Tokyo). 2000; 48: 1452-1457.
- Venkatadasu V, Muralidhar RV, Panda T. Analytical techniques for griseofulvin. Biop Eng. 2000; 22: 201-204.