Research Article
Austin J Biotechnol Bioeng. 2014;1(5): 5.
Genetic Variations among Ten Isolates of Fusarium Equiseti (Corda) Saccardo Isolated from Fruits and Vegetables
Bonde SR, Gade AK and Rai MK*
Department of Biotechnology, SGB Amravati University, India
*Corresponding author: Mahendra K Rai, Department of Biotechnology, S.G.B. Amravati University, Amravati - 444602 (Maharashtra), India.
Received: September 22, 2014; Accepted: November 01, 2014; Published: November 03, 2014
Abstract
We studied ten- isolate of F. equiseti isolated from different fruits and vegetables, which were identified on the basis of morphological characteristics and Random amplified polymorphic DNA (RAPD) analysis of these isolates was made by using 25 different universal decamer primers to confirm the identity. UPGMA study differentiation of ten isolates into two main clusters on the basis of similarity. The aim of the present study was to assess genetic variations among ten- isolate of F. equiseti by isolated from fruits and vegetables RAPD technique. The dendrogram obtained from the data showed that hierarchical clustering separated the isolates into three groups according to their similarity coefficients. The similarity coefficients among the all isolates ranged from 0.02 to 0.15.
Keywords: Dendrogram; Fusarium equiseti; Genetic diversity; RAPD; UPGMA
Introduction
The Fusarium species are commonly associated with many economically important crop diseases, such as, vascular- wilt, root-rot, stem- rot, and fruit and vegetable decay. The disease can cause yield and economic losses, hence study of distribution and diversity of these species is very important [1,2]. Nelson et al. [3] illustrated the morphology of F. equiseti. The fungus found to be associated with various diseases like cankers of sour cherry trees [4], rots of pumpkin [5] or cucurbit fruits in contact with soil [6]. It is resistant to antifungal agents like fluconazole and flucytosine [7].
Isolates of F. equiseti can be distinguished on the basis of growth rate and other morphological characters like shape and size of conidia on PDA [3]. However, molecular markers like random amplified polymorphic DNA (RAPD) can be used for confirmation of identity and differentiation among different isolates. RAPD assays have been used extensively to define fungal populations at species, intraspecific, race and strain levels. The use of molecular marker based on the polymerase chain reaction for species identification and as a diagnostic tool became very popular during the last decade [8] and usually RAPD-PCR technique is used for detecting genetic variability [9]. The morphological and molecular characterization of Fusarium species within the Liseola section isolated from corn grains were studied in different geographic regions of Brazil [10]. Moreover Fusarium wilt caused by F. oxysporum f.sp. melonis (FOM) phylogenetic analysis was performed by random amplified polymorphic DNA (RAPD) profiling and by translation elongation factor-1α (TEF-1α) sequencing [11]. Katkar et al., [12] reported F. oxysporum f.sp.ciceri were screened by using 30 RAPD primers for evaluation of genetic diversity. The biodiversity of Fusarium sps. isolated from the roots of oil palm and date palm in studied using RAPD molecular marker [12].
The aim of the present study was to assess the genetic variation among ten- isolates of F. equiseti (Corda) Saccardo isolated from fruits and vegetables by using RAPD-PCR method.
Materials and Methods
Isolation and morphological identification
Infected fruits were collected from different locations of Amravati city of Maharashtra (Table 1).
S.N.
Hosts
Botanical name
Place of collection
1
Brinjal (Fruit) (BB)
Solanum melongena
Amravati
2
Tomato (Fruit) (CC)
Lycospersicum esculentum
Amravati
3
Carrot (Stem) (DD)
Daucus carota
Amravati
4
Papaya(Fruit) (EE)
Carica papaya
Amravati
5
Papaya (Fruit) (FF)
Carica papaya
Amravati
6
Banana (Fruit) (GG)
Musa paradisiaca
Amravati
7
Beet ( Vegetable) (HH)
Beta vulgaris
Amravati
8
Potato (Tuber) (II)
Solanum tuberosum
Amravati
9
Ladies finger (Vegetable) (JJ)
Abelmoschus esculentus
Amravati
10
Papaya (Fruit) (KK)
Carica papaya
Amravati
Table 1: Fusarium equiseti isolated from different fruits and vegetables.
Different isolates of F. equiseti were recovered from infected materials on potato dextrose agar (PDA) and incubated at 25 ± 2°C for 3-4 days.
All the isolates of F. equiseti were identified on the basis of morphological and cultural characters [9].
DNA extraction
The total DNA was extracted using fungal genomic DNA isolation kit from Chromous Biotech Pvt. Ltd, Bangalore, India according to manufacturer's instructions.
RAPD analysis
The isolated DNA of ten different isolates of F. equiseti were screened using twenty- five random fungal primers (Table 2) from Random Fungal Primer Kit (RFu 'D'), Genie Pvt. Ltd, Bangalore, India, Amplifications were performed using PCR mixture in a total volume of 25 μL containing 12.5μl PCR master mix (2X) (Fermentas Life Sciences, Canada) 5 μl of template DNA (20 ng), 1.5 μl MgCl2 (25 mM), 0.3 μl Taq DNA polymerase (Genexy, 5U/μl), 1 μl each primer and 4.7 μl nuclease free distilled water (supplied with Fermentas PCR master mix).PCR was carried out on gradient PCR machine (Palm- Cycler Corbett Research, Australia). The programme included an initial denaturation at 94°C for 2 min, 35 cycles with denaturation at 94°C for 30 sec, annealing 40 °C for 1 min, extension at 72°C for 2 min and final extension at 72°C for 5 min with holding temperature at 4°C for 10 min. All experiments were repeated for three times. PCR products were electrophoresed on 1.5% agarose by using 1X TAE buffer (Fermentas Life Sciences, Canada), stained with ethidium bromide, visualized in a UV-transilluminator and the gels were photographed using Gel Doc (AlphaImager, Gel documentation system, USA), system.
S. N.
Primer
Sequences 5'-3'
No. polymeric bands
No. of monomeric bands
Total bands
%of plymorphism
1
RFu 1
CCTGGGCCAG
0
0
0
0
2
RFu 2
CCTGGGCGAG
0
0
0
0
3
RFu 3
CCTGGGCTGG
0
0
0
0
4
RFu 4
CCTGGGCTAT
0
0
0
0
5
RFu 5
CCTGGGCTTG
2
4
107
1.9
6
RFu 6
CCTGGGCTAC
0
0
0
0
7
RFu 7
CCTGGGCTTA
0
0
0
0
8
RFu 8
CCTGGGTCGA
0
0
0
0
9
RFu 9
CCTGGGTGCA
0
0
0
0
10
RFu 10
CCTGGGTGAC
2
2
90
2.2
11
RFu 11
CCTGGCTTAC
0
0
0
0
12
RFu 12
CCTGGGTTAC
0
0
0
0
13
RFu 13
CGGGGGATGG
0
0
0
0
14
RFu 14
CTCCCTGACC
0
0
0
0
15
RFu 15
GAGCACCTGT
0
0
0
0
16
RFu 16
GAGCACGTCA
0
0
0
0
17
RFu 17
GAGCACGGCA
0
0
0
0
18
RFu 18
GAGCACGGAG
0
0
0
0
19
RFu 19
GAGCTCGCAT
0
0
0
0
20
RFu 20
GAGGGCATGT
0
0
0
0
21
RFu 21
CCGGCCCCAA
0
0
0
0
22
RFu 22
CCGGCCTTAA
0
0
0
0
23
RFu 23
CCGGCCATAC
1
4
77
1.3
24
RFu 24
CCGGCCTTCC
0
0
0
0
25
RFu 25
CCGGCTGGAA
1
3
82
1.2
Table 2: Primer and their sequences used in RAPD-PCR test.
Results and Discussion
Morphological and cultural studies
All the ten- isolate of F. equiseti grew rapidly with dense aerial mycelia on potato dextrose agar. The detailed morphological studies including macroscopic and microscopic characteristics have been given in Table 3.
S.N.
Host
Colony Color
Type of Mycelium
Colony diameter*
(cm)
Common Conidial septation
Length of macroconidia** (µm)
Meso-conidia
Chlamydo-spore
1
Brinjal (Fruit) (BB)
White
Cream
Aerial
6.1
4-7
58.42 ± 1.71
Present
Present
2
Tomato (Fruit) (CC)
White
Brown
Aerial
8.1
3-6
47.82± 0.52
Absent
Present
3
Carrot (Stem) (DD)
White
Brown
Aerial
8.6
3-4
26.27± 0.16
Present
Absent
4
Papaya(Fruit) (EE)
Cream
Cream
Aerial
9.1
3-7
42.06 ± 0.15
Absent
Present
5
Papaya (Fruit) (FF)
Cream
Brown
Aerial
8.2
3-5
43.52 ± 3.31
Absent
Present
6
Banana (Fruit) (GG)
White
Cream
Aerial
8.2
3-6
56.41 ± 0.24
Absent
Present
7
Beet ( Vegetable) (HH)
Cream
Yellowish
Aerial
7.6
3-5
23.55 ± 0.13
Absent
Absent
8
Potato (Tuber)
White
Brown
Aerial
8.1
3-5
32.33 ± 1.40
Present
Present
9
Ladies finger (II) (Vegetable)
Cream
Brown
Aerial
8.2
3-4
34.2 ± 1.46
Present
Present
10
Papaya (JJ)
Yellow
Brown
Aerial
8.0
3-6
44.11 ± 0.44
Absent
Present
Table 3: Morphological characteristics of F. equiseti associated with different fruits and vegetables.
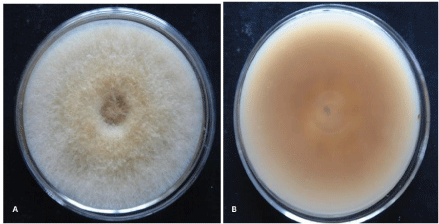
Macroscopic characteristics: Ten different isolates of F. equiseti were recovered from infected fruits and vegetables (Table 1) on Potato Dextrose Agar (PDA) and identified on the basis of morphological and cultural characteristics. The mycelium of F. equiseti was initially yellow but became brown with increasing age. It produces pale-brown to dark-brown pigments on PDA plate (Figure 1A, B). All the isolates exhibited similar growth rate of about 7.1-9 cm after 6 days incubation at 25 ± 20C. Morphological characters of F. equiseti showed resemblance with morphological characters reported by many researchers in their study carried out in the past for F. equiseti [2,3].
Figure 1: Growth of F. equiseti on PDA medium (A) Dorsal view (B) Ventral view isolated from Brinjal (Solanum melongena).
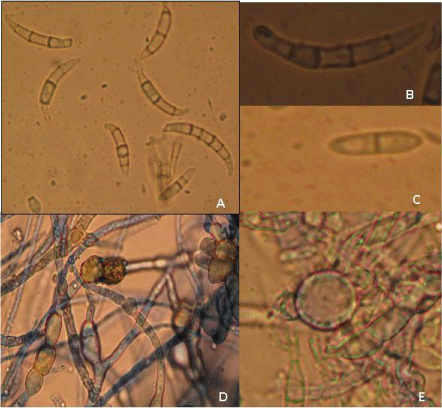
Microscopic characteristics: Microscopic examinations showed the presence of septate, hyaline hyphae and absence of microconidia in majority of isolates. Macroconidia were long slender, abundant and borne on aerial mycelium, generally 5-7 septate and 23.55 to 58.45 μm in length (Figure 2A). The apical shape of macroconidia was tapered and elongated or even whip-like while the basal shape was prominent foot-shaped (Figure 2B). Bi-septate mesoconidia were found in a few isolates (Figure 2C). Two types of chlamydospores were found in the present study. Interestingly, in one case chlamydospores were found in the hyphae singly and sometimes in chains (Figure 2D), while in another case, single chlamydospores were present. (Figure 2E) which is a specific characteristic of F. equiseti. It provides the strength for the identification of all these isolates on the basis of microscopic characteristics. Aigbe et al.[2] reported the similar microscopic characters for F. equiseti isolated from cow pea in Nigeria.
Figure 2: Microscopic characteristics of F. equiseti (A) Macroconidia (B) Single Macroconidium (6 septate) (tapering apical and notched foot shape) (C) Mesoconidia (D) Chlamydospores in chain in hyphae (E) Chlamydospore singly in conidia.
RAPD analysis
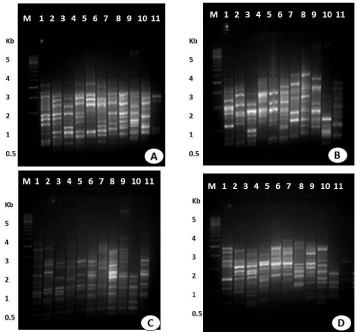
RAPD-PCR analysis were carried out to screen genomic DNA isolated from ten isolates of F. equiseti using twenty five random decamer primers of Fungal RAPD Primer (RFu 'D') kit (Table 2). The culture of F. equiseti procured from IMTECH (MTCC 3731), Chandigarh was taken as reference for RAPD study. In the preliminary experiments, twelve out of the twenty five primers tested produced distinct and reproducible band profile, and polymorphisms produced by ten primers. Four of twelve primers were used for comparative analysis of ten isolate of F. equiseti. The primers, including RFu 5 (5'-CCTGGGCTTG-3'), RFu 10 (5'-CCTGGGTGAC-3'), RFu 23 (5'-CCGGCCATAC-3'), and RFu 25 (5'-CCGGCTGGAA-3') generated polymorphic bands in all ten isolates of F. equiseti (Figure 3). Primers RFu 5, RFu 10, Rfu 23 and RFu 25 showed monomorpic and polymorphic bands (Table 2). The amplified fragments were ranged 0.5 kb to 3.0 kb, except a single band which ranged upto 5.0 kb. RAPD assays of all ten isolates with four above mentioned primers yielded 170 bands, which were found to be polymorphic. Above data showed that RAPD is a convenient method for distinguishing the isolates of F. euiseti and also reveal a significant genetic variation among these isolates. F. oxysporum also demonstrated the genetic variation and differentiation among its isolates. Assigbetse et al. [14] differentiated races of F. oxysporum f. sp. vasinfectum on cotton by using RAPD as a molecular tool. They reported the significant genetic variation in these isolates of F. oxysporum. In another study, Edel and colleagues [15] observed that the soil isolates of F. oxysporum in France showed genetic diversity. Our finding supports the work carried out by Leslie and co-workers [9]. They observed inter- and intra specific genetic variation in different Fusarium species. RAPD-PCR technique was used to rapid identification and differentiation of Fusarium species [16]. Moreover Costano [17] studied genetic diversity of F. oxysporum f. sp. dianthi in Southern Spain in 132 isolates collected from carnation wilted plants. RAPD marker used to estimate genetic variation among 12 isolates of the F. solani isolates causing dry root rot of sweet orange (Citrus sinensis osbeck) [18].
Figure 3: RAPD patterns on 1.5% agarose gel of amplified fragments generated from 10 isolates of F. equiseti with random primers (A) RF u-5 (B) RFu - 10 (C) RFu-23 (D) RFu-25. Lane M- DNA marker (100 bp), lane 1- mtcc- 3731, lane 2- Brinjal (Solanum melongena), lane 3- Tomato (Lycospersicum esculentum), lane 4-Carrot (Daucus carota), lane 5-Papaya 1 (Carica papaya), lane 6-Papaya 2 (Carica papaya), lane 7-Banana (Musa paradisiaca), lane 8- Beet (Beta vulgaris), lane 9-Potato (Solanum tuberosum), lane 10-Ladies finger (Abelmoschus esculentus), lane 11-Papaya 3 (Carica papaya).
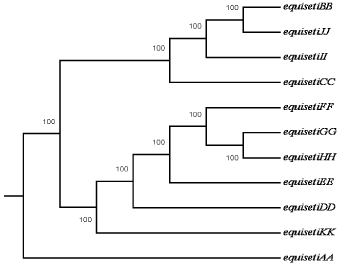
Dendrogram showing genetic relationship among the ten isolates of F. equiseti
AA) MTCC 3731, (BB) Brinjal (Solanum melongena), (CC) Tomato (Lycospersicum esculentum), (DD) Carrot (Daucus carota), (EE) Papaya 1 (Carica papaya), (FF) Papaya 2 (Carica papaya), (GG) Banana (Musa paradisiaca), (HH) Beet (Beta vulgaris), (II) Potato (Solanum tuberosum), (JJ) Ladies finger (Abelmoschus esculentus), (KK) Papaya 3 (Carica papaya).Figure 4:Dendrogram showing genetic relationship among the ten isolates of F. equiseti
AA) MTCC 3731, (BB) Brinjal (Solanum melongena), (CC) Tomato (Lycospersicum esculentum), (DD) Carrot (Daucus carota), (EE) Papaya 1 (Carica papaya), (FF) Papaya 2 (Carica papaya), (GG) Banana (Musa paradisiaca), (HH) Beet (Beta vulgaris), (II) Potato (Solanum tuberosum), (JJ) Ladies finger (Abelmoschus esculentus), (KK) Papaya 3 (Carica papaya).
Unweighted Pair Group Method with Arithmetic Mean analysis (UPGMA) Cluster Analysis
F. equiseti isolates were analyzed using UPGMA clustering approach with bootstrap value of 1000. UPGMA, a multivariate statistics analysis of the RAPD data separated the F. equiseti in two clusters. UPGMA dendrogram showed MTCC (3731) culture of F. equiseti and other isolates reported from India in two different clades. F. equiseti isolated from brinjal (BB) (Solanum melongena), tomato (CC) (Lycospersicum esculentum), potato (II) (Solanum tuberosum) and ladies finger (JJ) (Abelmoschus esculentus) were in one clade while other isolates were in different clades. Isolates of Fusarium recovered from brinjal, potato and ladies finger were similar but F. equiseti isolated from tomato showed genetic variation as it was present in different clade in dendrogram. UPGMA analysis thus carried out in the present study showed the genetic variation in these isolates of F. euiseti. A distance matrix on simple matching coefficients was calculated from the data based on the RAPD of all ten isolates of F. equiseti. The matrix was used to construct a dendrogram using distance tool with UPGMA method of PHYLIP for establishing to analyze the level of relatedness among the ten isolates. The dendrogram obtained from the data showed that hierarchical clustering separated the isolates into three groups according to their similarity coefficients. The similarity coefficients among the all isolates ranged from 0.02 to 0.15. Distance matrix of different isolates of F. equiseti was analyzed (Table 4).
Isolates
MTCC 3731
Brinjal
Tomato
Carrot
Papaya 1
Papaya-2
Banana
Beet
Potato
Ladies Finger
Papaya 3
MTCC 3731
0.000
Brinjal
0.050
0.000
Tomato
0.066
0.031
0.000
Carrot
0.074
0.042
0.043
0.000
Papaya 1
0.063
0.038
0.045
0.051
0.000
Papaya 2
0.074
0.060
0.089
0.057
0.031
0.000
Banana
0.051
0.036
0.060
0.031
0.034
0.019
0.000
Beet
0.052
0.043
0.060
0.045
0.051
0.036
0.016
0.000
Potato
0.040
0.031
0.051
0.074
0.052
0.063
0.041
0.029
0.000
Ladies Finger
0.047
0.020
0.034
0.062
0.051
0.074
0.055
0.046
0.021
0.000
Papaya 3
0.110
0.089
0.132
0.156
0.087
0.146
0.156
0.159
0.082
0.083
0.000
Table 4: Distance matrix by UPGMA Analysis.
UPGMA is a simple agglomerative or hierarchical clustering method used in bioinformatics for the phylogenetic analysis. The results obtained in the present study are noteworthy and showed the similarity with other researchers. In their studies on isolates of F. semitectum, F. solani and F. oxysporum respectively, they used data generated from RAPD banding pattern for the UPGMA analysis and found that there were genetic variations in different isolates of same Fusarium [19-21]. A modified mathematical model by Nei and Li [22] for the evolutionary change of restriction site was used. Although the ten isolates of F. equiseti were recovered from different infected fruits and vegetables showed the similar morphological characteristics. Interestingly, our results suggest a significant genetic variation among the isolates of F. equiseti. It seems that the fungal species showed the host dependent genetic variation. Similarly biodiversity of sixteen Fusarium isolates, isolated from the roots of oil palm and date palm in Nigeria was studied. In this RAPD was used to detect the phylogenetic similarity between them. The UPGMA dendrogram clearly separated these sixteen Fusarium isolates into five groups (clusters). The first at SC values of 100 grouped six Fusarium isolates of both oil and date palms [13].
Conclusion
Our results suggest a significant genetic variation among the isolates of F. equiseti by RAPD analysis, the genetic variation shown by fungal species is host dependent. We report for RAPD analysis for genetic variation among different isolates of F. equiseti isolated from fruits and vegetables. We suggest that RAPD marker may be used as one of the reliable alternatives for the determination of genetic variation among the Fusarium species when a little variation in morphological characters exist.
Acknowledgement
The authors are grateful to Defense Research and Development Organization (DRDO), New Delhi for providing financial assistance for the present research.
References
- Latiffah Z, Zariman M, Baharuddin S. Diversity of Fusarium species in cultivated soils in Penang. Malaysian Journal of Microbiology. 2007; 3: 27-30.
- Aigbe SO, Fawole BA. Cowpea Seed Rot Disease Caused by Fusarium equiseti Identified in Nigeria. ASP Net. 1999; 83: 9641.
- Nelson PE, Toussoun TA, Marasas WF. Fusarium species: an illustrated manual for identification. The Pennsylvania State University Press, University Park. 1983.
- Olszak M. Aetiology of sour cherry fungal diseases in Poland III pathogenicity of the isolated fungi. Journal of Fruit and Ornamental Plant Research. 1994; 2:165-184.
- Elmer WH. Fusarium fruit rot of pumpkin in Connecticut. Plant Disease. 1996; 80:131-135.
- Adams GC, Gubler WD, Grogan RG. Seedling disease of muskmelon and mixed melons in California, USA caused by Fusarium equiseti. Plant Disease.1987; 71:370-374.
- Pujol I, Guarro J, Gené J, Sala J. In-vitro antifungal susceptibility of clinical and environmental Fusarium spp. strains. J Antimicrob Chemother. 1997; 39: 163-167.
- Ingle A, Karwa A, Rai M. K, Gherbawy Y. Fusarium: Molecular detection, mycotoxins and biocontrol. In: Gherbawy Y, Mach R, Rai M. editors. Current Advances in Molecular Mycology, Science Publishers Inc., Enfield, New Hampshire 03748, USA. 2009; 85-106.
- Leslie JF, Summerell BA. The Fusarium laboratory manual, 3rd ed. Blackwell publishing professional, Ames, IA, USA. 2006; 168-169.
- Lanza FE, Zambolim L, da Costa R, Queiroz V, Cota J, da Silva D, et al. Prevalence of fumonisin-producing Fusarium species in Brazilian corn grains. Crop Protection. 2014; 65:232-237.
- Katkar M, Mane SS. Characterization of Indian races of Fusarium oxysporum f.sp. ciceri through RAPD markers. International Journal of Agriculture, Environment and Biotechnology. 2014; 5: 323-328.
- Singh PK, Sharma H, Shrivistava N, Bhagyawant S. Analysis of Genetic Diversity among Wild and Cultivated Chickpea Genotypes Employing ISSR and RAPD. American Journal of Plant Sciences. 2014; 5: 676-682.
- Chidi N I, Adekunle AA, Eziashi EI, Omamor IB, Odigie EE, Osagie IJ. Molecular Identification of Biodiversity of Fusarium species Isolated from Wilted Oil Palm and Date Palm in Nigeria. British Journal of Biotechnology. 2014; 4: 612-621.
- Assigbetse KB, Fernandez D, Dubois MP, Geiger JP. Differentiation of Fusarium oxysporum f. sp. vasinfectum races on cotton by Random amplified polymorphic DNA (RAPD) analysis. Phytopathology. 1994; 84: 622-626.
- Edel V, Steinberg C, Gautheron N, Recorbet G, Alabouvette C. Genetic diversity of Fusarium oxysporum populations isolated from different soils in France. FEMS Microbiol Ecol. 2001; 36: 61-71.
- El-Fadly GB, El-Kazzaz M, Hassan M, El-Kot GA. Identification of some Fusarium spp. using RAPD-PCR technique. Egyptian Journal Phytopathology. 2008; 36: 71-80.
- Castaño R, Scherm B, Avilés M. Genetic Diversity of Fusarium oxysporum f. sp. dianthi in Southern Spain. Jounal of Mycology. 2014; 1-14.
- Sankar TG, Gopal K, Gopi V Sreenivasulu Y. Molecular characterization of Fusarium solani isolates causing dry root rot of sweet orange (Citrus sinensis osbeck). International Journal of Current Microbiology and Applied Sciences. 2014; 3: 105-114.
- Abd-Elsalam KA, Schnieder F, Asran-Amal A, Khalil MS, Verreet JA. Intra-species genomic groups in Fusarium semitectum and their correlation with origin and cultural characteristics. Journal Plant Disease Protection. 2003; 110: 409-418.
- Ingle AP, Rai MK. Genetic diversity among Indian phytopathogenic isolates of Fusarium semitectum Berkeley and Ravenel. Advances in Bioscience and Biotechnology. 2011; 2:142-148.
- Bonde SR, Gade A, Rai M. Genetic Diversity among Fourteen Different Fusarium Spp. Using RAPD Marker, Biodiversitae. 2013; 14: 55-60.
- Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979; 76: 5269-5273.