
Research Article
Austin J Biotechnol Bioeng. 2023; 10(2): 1125.
Molecular Characterization of SHV, BLA-OXA and B-Lactamase Resistant Genes Leading to Computational Based Inhibitor Screening Against Bacillus Species
Nida Khan1; Eisha Munaf2; Muhammad Asif Muneer3*; Muhammad Hamza Ashraf3; Ayesha Mumtaz3; Muhammad Zeeshan4; Kamran Khan5; Muhammad Rizwan Ali6
1Department of Biotechnology, University of Central Punjab, Lahore, Pakistan
2Department of Biotechnology, Jinnah University for Women, Karachi, Pakistan
3Centre for Applied Molecular Biology, University of the Punjab Lahore, Pakistan
4Insitute of Microbiology and Molecular Genetics, University of the Punjab Lahore, Pakistan
5Shaheed Benazir Bhutto University Dir Upper, Pakistan
6Centre of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan
*Corresponding author: Muhammad Asif Muneer Centre for Applied Molecular Biology, University of the Punjab Lahore, Pakistan. Email: axifmalick786@gmail.com
Received: October 25, 2023 Accepted: November 27, 2023 Published: December 04, 2023
Abstract
Antibiotic resistance in bacterial pathogen is a great challenge that is connected with excessive morbidity and mortality of living beings. A common motive of antibiotic resistance in bacteria is an increased abundance of Β- lactamases. There chromosomal Β- lactamases do not generally provide effective antibiotic resistance in wild type Bacilli despite evidence that the genes are not completely silenced. Under antibiotic selection pressure however, a number of strains show increased resistance suggesting mutation induced up regulation of Β- lactamases. Computer-assisted drug design has made significant progress in predicting biologically active molecules and their receptor binding conformation, with results that are sometimes more exact than those acquired through traditional methods like as high-throughput screening. The results of computational studies are sometimes more precise than the experimental possibilities, and they contribute to the improvement of the experimental array. In this study, identified pathogenic species of bacillus, and analyzed the antibiotic resistant genes. The Previous study of its characterization showed the presence of BLA-OXA and SHV genes in Bacillus cereus and Bacillus paramycoides. The sequenced antibiotics resistant genes were subjected to computational analysis for the interpretation of the potential inhibitors against BLA-OXA and SHV. The analyzed inhibitor will enable to study the diversity of antibiotic resistant mechanism and to minimize the resistance of bacterial species against antibiotics.
Keywords: Antibiotic Resistance; B- lactamases; Pathogenic species; Computational Analysis; Horizenal gene transfer; Molecular docking
Introduction
Antibiotic resistance in bacterial strains is being produced by an increase in the quantity of beta-lactamases. Many bacterial strains, including Bacillus bacterial species, include beta-lactamase resistance genes in their wild-type genomes. Their chromosomal beta-lactamase do not offer efficient antibiotic resistance in the wild-type of strain, but the evidence of the genes is completely muted. In the face of antibiotic selection pressure, a large number of bacteria with abundant resistance may undergo mutation-induced upregulation of beta-lactamase production [1]. The commercialization and administration of antimicrobial agents against infection changed treatment patterns following the revolutionization of contemporary medicines. Antibiotics have become key medications in medical procedures such as surgical techniques, cancer therapy and organ transplantation, and other therapies employing with many other techniques [2]. However, the negative consequence is in the importance of the establishment of resistance in bacterial strains, which is currently jeopardizing treatment successes and patient success [3]. In 1927, a scientist called Sir Alexander Fleming developed penicillin, the first antibiotic. Antibiotic resistance research began in 1940 with the study of microbes. A few years ago, the beta-lactam category of antibiotics accounted for over 60% of all antibiotics used in human and animal therapeutics that are resistant to gram-negative bacteria [4]. Beta-lactams kill germs by inhibiting cell wall formation. A protein known as beta lactamase will have hydrolyzed the b-lactam ring, allowing the bacteria to live [5]. Currently, 70% of commercially available antibiotics are ineffective against pathogenic bacteria, causing serious problems in general health concerns of public concern. Antibiotic resistance is the World Health Organization's (WHO) third major public health concern. Antibiotic resistance is a major scientific problem in hospitals and communities [6]. Fast identification of antibiotic-resistant organisms in clinical laboratories is required to identify judicious antibiotic-defiance bacterial species [7].
The creation of expanded range lactamases (ESBLs) is a key obstruction system that defers anti-toxin treatment of illnesses. These contaminations are brought about by Enterobacteriaceae and represent a significant danger to the anti-microbial munitions stockpile at present accessible [8]. ESBLs are isolated into bunches in light of their amino corrosive grouping homology. Notwithstanding, legitimate disease control measures and hindrances are expected to forestall the spread and breakout of microscopic organisms that produce ESBL [9]. The two portions within the chemical or that attacked its own molecule are the most important strategies used by bacteria to manage the presence of antibiotics. As a result, the antibiotic fails to engage with its target [10]. The major mechanism of Β-lactam resistance has been the elimination of these chemicals by the activity of an enzyme known as Β-lactamases. These enzymes break the chemical link, i.e. the amide bond, present in the -lactam ring, rendering the antibiotic useless [11].
Medicines of organism diseases with antimicrobial meds are ineffectual inferable from multidrug opposition; this issue is definitely not restricted to microorganisms yet additionally to microorganisms that can possibly advance and render new medications inadequate [12]. A few bacterial strains are impervious to a solitary anti-microbial, while many are impervious to many medications, coming about in multidrug safe bacterial strains or superbugs. With the disclosure of anti-toxins, for example, sulfonamides and penicillin during the 1940s, another time of working on human wellbeing and relieving diseases began, yet with it came the development of anti-microbial opposition [13]. During the 1950s-1970s, the "golden period" of antibiotic research, widespread the use of medications led to the development of numerous new antibiotic classes. Unfortunately, microorganisms were shown to be resistant to every newly discovered medicine during clinical trials several years following their development [14]. As the number of Multi-Drug Resistant Organisms (MDROs) grows, the efficiency of our current antibiotics will deteriorate. Antibiotic resistance is accelerated by the widespread use of antibiotics and the spread of antibiotic-resistant genes in the bacterial population [15].
As a result, antibiotics are rendered inactive by the activity of beta-lactamase enzymes, which are generated in bacteria and can impair the antibiotic's ability to bind to a target. Beta lactamase degrades the amide bonds found in -lactam antibiotics, rendering them useless against infections. Broad spectrum antibiotics include Penam, Penicillins, and its derivatives such as Oxacillin, Methicillin, and Cephalosporins [16]. Serratia marcescens and numerous bacillus strains, including Bacillus cereus and Bacillus paramycoides, showed total aversion to meropenem (100 percent), dictionary (100 percent), ciprofloxacin (90.5%), and gentamicin (84.6%), as well as outright (100 percent) protection from ceftazidime, cefuroxime, ampicillin, and Augmentin. All of the disengages (100 percent) created ESBLs, albeit just 33.3% delivered carbapenems (Zeng and Lin, 2013). The beta lactamase opposition qualities Ampicillins (AmpC), cefoxitin variety (FOX-1) ampicillins, cefotaxime (CTX-M), Sulfhydryl Variable (SHV), and Temoniera (TEM) were found in 15.4 percent, 0.0 percent, 53.9 percent, 38.5 percent, and 15.4 percent of Serratia marcescens, individually. Essentially, co-carriage of two and three distinct obstruction qualities was seen as in four (30.8 percent) and one (7.7 percent) secludes, separately [17]. Carbapenems beta-lactamase molecules are classified into three classes: A, B, and D. Acinetobacter baumannii mostly contains Class D Oxacillinases (OXA type), whereas Pseudomonas aeruginosa primarily contains Class B (Metallo-Β-lactamases).There are two types of antibiotics: bactericidal medications that induce cell death and bacteriostatic drugs that just limit cell growth [18]. Antibacterial drugs now on the market involves the interference in the formation of the Nucleic Acids and cell wall, inhibition of Protein synthesis, metabolic pathways, membrane functions and ATP synthase. Antibiotic-induced cell death is triggered by a physical contact between a particular target of bacteria and medication molecules. It does, however, comprise a complicated process that involves changes in the afflicted bacteria at several levels, such as molecular and biochemical [19].
Antibiotic Resistance
Sir Alexander Fleming, a scientist, created the most important antibiotic, penicillin, in 1927. Antibiotic resistance research began in 1940 with the study of microbes. A few years ago, the beta-lactam category of antibiotics accounted for over 60% of all antibiotics used in human and animal therapeutics that are resistant to gram-negative bacteria [20]. Beta-lactams kill germs by inhibiting cell wall formation. The beta-lactam ring is dissolved by a protein called beta lactamase, which allows the bacteria to live. Which gradually grow, and eventually, even subsequent generations of various antibiotics may become disabled [16].
As a result, in the 1980s, Gram-Negative (GN) rods were discovered to have an enzyme spectrum known as Extended-Spectrum Β-Lactamases (ESBLs). Because ESBLs are kept on plasmids, they are easily transferred to new bacteria. Microbes that are resistant to beta-lactam antibiotics have a light-emitting diode that activates in the presence of other antibiotics such as aminoglycosides and fluoroquinolones [22]. These antibiotics have a halting effect on bacterial proteins or DNA production. At the same time, resistance to this category of medicines spreads throughout numerous germs, resulting in many bacteria being labelled as Multi-Drug Resistant (MDR) [23]. Antibiotic resistance has been designated as the third significant public health problem in the twenty-first century by the World Health Organization (WHO). Resistance to antibiotics is major factual examination in the two hospitals and communities. Fast detection in clinical laboratory is required to identify judicious antibiotic resistance organisms [2]. The generation of ESBLs is an essential mechanism of resistance which slows antibiotic illness therapeutics. These septicemias are attributed to Enterobacteriaceae, as well as they pose substantial danger in order to antibiotic arsenal now assessable. ESBLs are divided against different classes based on the amino acid classification of similarity. To prevent the spread and breakout of bacteria that produce ESBL, effective infection control techniques and barriers are essential [24]. Antibacterial defiance enables the capability of bacteria to resist the ramification against germicides, which are drugs that kill or slow the growth of germs such as penicillin or ciprofloxacin. When bacteria grow resistant to antibiotics, treating diseases caused by those germs becomes more difficult and treatment options become limited [25]. Anti-infection opposition can damage individual's biological system at distinctive stages in life, as well as the medical services, veterinary, and agribusiness ventures. Subsequently, it is one of the world's most squeezing general medical problems. In the United States, more than 2.8 million anti-infection safe contaminations happen every year. As indicated by the CDC's 2019 Antibiotic Resistance (AR) Threats Report, in excess of 35,000 individuals die consequently [26]. Irresistible illnesses are a main source of dreariness and mortality in the globe today. Lower respiratory contamination, diarrheal ailments, HIV/AIDS, and intestinal sickness are among the best 10 supporters of dismalness and mortality, as indicated by a WHO appraisal of these illnesses. Antimicrobial opposition has extensively expanded the impact of irresistible illnesses, as well as the quantity of diseases and medical services costs [27]. Notwithstanding the way that we have a tremendous assortment of antimicrobial specialists from which to decide for potential disease treatment, antimicrobial opposition has been recorded for every one of them, and obstruction grows rapidly once another anti-infection is endorsed for utilization. The World Health Organization (WHO) sent off a Global Action Plan on Antimicrobial Resistance in 2015 because of these worries [28].
Beta Lactams Antibiotics
Antibiotics classed as beta lactams are a large range of antibacterial medicines defined by their chemical structures. Gram negative rods, with the exception of carbapenem, are able to withstand the lethal effects of these medications due to their ability to produce -lactamase, an enzyme that hydrolyzes all -lactam antibiotics [29]. These bacteria cause an increase in carbapenem drug demand, resulting in resistance gene mutation in the bacteria. This mutation results in the creation of the most dreaded -lactamase enzyme (Carbapenemase), which can hydrolyze all -lactam medicines as well as carbapenem [30].
Carbapenemase Producers
All -lactam antibiotics, such as carbapenem, penicillins, and Cephalosporin are rendered inactive by carbapenemase manufacturers. In GNB, resistance is caused by two mechanisms: first, the acquisition of Carbapenemas encoding genes, which encode Carbapenemase, which degrades beta lactam; and second, the acquisition of Carbapenemase encoding genes, which encode Carbapenemase, which degrades Β-lactam [31]. Second, decreased protein expression, either qualitatively or quantitatively, reduces antibiotic absorption, as does overexpression of the -lactamase enzyme, which has a weak interaction with Carbapenems. Carbapenemase manufacturers fall into the following categories [31].
Carbapenemase Class A
NmcA, Sme, IMI-1, and SFC-1 are chromosomal encoded genes, while KPC, IMI-2, and GES are plasmid intermediated genes. KPC is the most frequent enzyme among these, and it has sparked popular interest [32].
Metallo Β-lactamase Class B
Impenemase (IMP), New Delhi Metallo-lactamase (NDM), and Veron-encoded Metallo-lactamase make up Class B Metallo-lactamase (VIM). Except for aztreonum, metallo lactamase makers can hydrolyze all -lactams. Ethyl diamine tetra acetic acid can impede their action in the lab (EDTA). Metallo-lactamases are classified as class B because they require zinc divalent cations as cofactors for enzymatic activity, and so are classified by metal chelator inhibition [33]. They show effective hydrolyzing activity against carbapenem and other -lactam antibiotics. Carbapenems, primarily imipenem, meropenem, and panipenem (available only in Japan), are effective agents for treating infections caused by multidrug-resistant Bacillus species. However, the number of carbapenem-resistant Pseudomonas aeruginosa strains has lately increased. MBL-producing Bacillus strains have therefore been identified, and they are key causes of nosocomial infections linked to clonal transmission [34].
Carbapenemase Class D
The OXA-48 strain was initially identified in Turkey in 2003 and is frequently isolated as the source of nosocomial outbreaks. This enzyme can now be found all over the world, especially in Europe (southern and eastern Mediterranean Sea) and Africa. While Queen A.M. reported a global expansion of CP along with an increase in KPC endemicity in the southern United States and Greece. Southern Europe and Asia have a high prevalence of metallo enzymes (VIM), whereas oxacillinase-48 has been found predominantly in the Mediterranean, European countries, and India and Pakistan [35].
Circumventing Β-Lactamase
When some drugs that block the active site, such as beta-lactams, are used, beta-lactamase-mediated resistance is diminished. This can be accomplished in a way by creating irreversible "suicide inhibitors", which are permanently inactivate the beta lactamase active site. Tazobactum, Sulbactam, and Clavulanic acid are examples of commercially available inhibitors in this category [36].
Amalgamation of Β-Lactam with Β-Lactam Inhibitors
PBPs are not generally inactivated by inhibitors, but there are a few notable exceptions: (i) sulbactam has intrinsic antibacterial activity against Acinetobacter spp., N. gonorrhoeae, and Bacteroides spp.; (ii) clavulanate influences N. gonorrhoeae and Haemophilus influenza. The antibacterial activity of all of these inhibitors, which are frequently recommended in conjunction with beta lactam antibiotics, is quite poor [37]. There are presently five beta lactam anti-microbial and beta lactam inhibitor combos accessible. In the United States, the anti-infection agents’ ticarcillin-clavulanate, amoxicillin-clavulanate, ampicillin mix with sulbactam, and piperacillin mix with tazobactam are clinically used. Cefetrizole related to sulbactam is generally utilized in Japan, India, and other European nations, yet not in the United States [38].
Amoxicillin-Clavulanate
These were first invented as well as made accessible for clinical use in the United States in 1981 and 1984, respectively. Clavulanate has no effect on the efficacy of amoxicillin in opposition to certain vulnerable microbes for example, S. aureus, Enterococci and E. coli. The addition of clavulanate to amoxicillin broadens its action spectrum in opposition of penicillinase-creating S. aureus, Moraxella Catarrhalis H. influenzae, N. gonorrhoeae, Bacteroides spp., E. coli, and Klebsiella spp. (Zhang et al., 2021).Amoxicillin-capacity clavulanate's to be controlled orally makes it ideal for use in short term centers, and this lactam-lactamase inhibitor mix essentially affects the treatment of local area procured respiratory contaminations [16].
Piperacillin-Tazobactam
Piperacillin has bactericidal effect against bacteria in the gramme negative category. In 1993, Piperacillin and Tazobactam were released in the United States. Piperacillin has bactericidal activity against streptococci, Pseudomonas aeruginosa, pneumococci, and Enterococcus faecalis as a single agent, and a combination of piperacillin with Tazobactam has this activity as well [39]. Combining Tazobactam with gram positive and other gram-negative bacteria which codes for AmpC genes does not appear to be successful. Tazobactam, then again, builds piperacillin's useful action in contrast to Enterobacteriaceae, bringing down MICs which produce ESBLs [40].
Ampicillin-Sulbactam
Ampicillin is an antibiotic that works well against both beta lactamase-producing and non-producing bacteria. When ampicillin was mixed with sulbactam, its activity rose (Shafiee et al., 2021). The mixture of ampicillin and sulbactam is utilized to analyze polymicrobial contaminations like gynecological and stomach a medical procedure disease, among others. Unfortunately, resistance to ampicillin-sulbactam has been observed in some E. coli isolates [41].
Mechanism of Antibiotic Resistance
A single bacterial stain can adopt only a few types of resistance mechanisms. Because some bacterial species are intrinsically resistant to antibiotics, resistance is determined by the type of the drug. Antibacterial insubordination is delegated either inborn or gained. Inborn opposition is intrinsic to a bacterial animal types and can't be given to different microscopic organisms, though gained obstruction is sorted into two kinds: biochemical and genetic [42]. Biochemical opposition incorporates instruments, for example, anti-microbial inactivation by enzymatic corruption, anti-microbial powerlessness to join to its predetermined objective because of an adjustment of target site, any decrease in drug fixation, and evolving titer. In terms of genetic mechanisms, bacteria can undergo mutation or parallel transfer [43]. The basic routes by which antibiotic resistant genes spread. The main mechanisms of gene transfer could be conjugation, transformation, and transduction. Antibiotic resistance genes can be found on plasmids and transposons, and conjugation is the most prevalent parallel gene mechanism, which involves the direct interaction of the bacteria, both the recipient and donor. Other route for gene transferring is a process by which a bacterial that enters a bacterial cell takes naked DNA from the environment, must be free of nuclease damage, and then incorporates into bacterial DNA [42]. Antibiotic resistance does not emerge as a result of transformation because numerous processes prevent the incoming DNA from surviving and integrating. Because heat, chemical degradation, and nuclease action can destroy DNA physically, it is difficult for DNA in various biological systems [44]. This process has been seen in organic product juices and different food items, but assuming food media are complicated, this mind boggling climate can safeguard DNA. In transduction, a bacteriophage connects to a bacterial cell and infuses its DNA, which is then consolidated into the bacterial DNA, bringing about new phage duplicates. Because bacteriophages are host specific, transduction occurs between closely related species [45].
Transfer of Antibiotic Resistant Genes
Once bacteria gain genes of antibacterial defiance, they become the absolute component of the bacterial DNA, which cannot be eliminated by any cost. Many GRAS (generally regarded as safe) lactobacilli species may act as a carrier for antibiotic resistance genes. These microbes are normally consumed in huge amounts and are in nearness to different microorganisms in the human gastrointestinal parcel, giving ideal conditions to even exchange of conjugative plasmids and transposons holding onto antimicrobial opposition qualities [46]. "Commensal bacteria in the colon, both those that potentially behave as opportunistic pathogens and those that are actually non-pathogenic, exchange DNA with one another," according to the "Resistance gene reservoir hypothesis" [47].
Horizontal Gene Transfer
Lactobacilli, which operate as vectors, can transfer antibiotic resistance genes. These bacteria are highly eaten and near to other bacteria in the human GIT, providing ideal circumstances for horizontal gene transfer that leads to antibiotic resistance [48].
Bacillus Species
Bacillus cereus is a poison delivering gram-positive bacterium that can be found in soil, vegetation, and food. It as often as possible causes queasiness, regurgitating, and loose bowels in the digestive organs. Nonetheless, in immunocompromised hosts, it has been connected to huge diseases, including septicemia and endophthalmitis, which can bring about vision misfortune [49]. B. cereus food contamination is an intense inebriation that happens when this microorganism makes poisons, bringing about one of two sorts of gastrointestinal sickness: emetic (regurgitating) or diarrheal disorder. B. cereus is a responably pervasive reason for gastroenteritis over the world. In 2006, it was assessed that around 36,000 occasions of B. cereus-related food contamination happened in Canada [50].
Diseases Caused by Bacillus Spp.
Bacillus cereus is a foodborne microorganism that produces poisons and can cause two sorts of gastrointestinal sickness: emetic (spewing) and diarrheal disorders. Retching happens following admission of debased food if the emetic poison (cereulide) is made in the food. Following admission of B. cereus-polluted food, enterotoxins are created in the digestive tract, bringing about the runs [51]. Purchasers of food debased with the emetic poison cereulide will get emetic condition; subsequently, the food should be sullied with B. cereus strains equipped for creating this poison and dealt with in a manner that works with bacterial development and ensuing poison age. B. cereus levels need be more prominent than 10,000 for every gram of food to make enough cereulide to cause heaving, but various articles have portrayed infection, including hospitalizations, with lower numbers [52]. Since the poison is made in the food and is heat safe, it won't be annihilated by most cooking strategies, in any event, when the vegetative cells have been inactivated. This condition is usually connected to bland food sources like pasta or rice dishes [53]. Whenever countless B. cereus vegetative cells (something like 10,000 for each gram of food) are devoured, enterotoxin is created in the small digestive system, causing loose bowels. Meat items, stews, soups, sauces, vegetables, and milk items have all been related to the diarrheal disease [53].
Antibiotic Resistance in Bacillus Spp.
B.cereus produces beta-lactamases, not at all like virtually all B.anthracis confines, it is impervious to beta-lactam antimicrobial medications, including third-age cephalosporins. Aminoglycosides, clindamycin, vancomycin, chloramphenicol, and erythromycin are typically viable against it. Visual diseases can be extremely perilous; visual deficiency can result in just 12 to 18 hours, with enucleation and visual impairment [54]. Assuming that capacity is to be safeguarded, brief antimicrobial treatment with fundamental, skin, and maybe intravitreal anti-infection agents should start before culture discoveries are accessible. Clindamycin in addition to gentamicin and vancomycin alone have both been demonstrated to be compelling, and imipenem might be advantageous also. To guarantee suitable antibacterial fixations in various region of the eye, numerous treatment pathways are required [55]. Skin clindamycin, for instance, has a high watery to glassy humor drug proportion since it conveys a high medication portion to the foremost compartment of the eye since amino glycosides are disposed of from the eye by a front course. This could clarify why central end ophthalmitis and those influencing the front section have a preferred guess over those influencing the back portion, which oftentimes bring about visual impairment. In spite of the way that beta-lactams specially disseminate into the glassy humor, the 3-lactamases delivered by B.cereus make them inadequate and improper for treating B.cereus diseases [56]. Be that as it may, there has been minimal definitive review on drug entrance into the aggravated eye, and most treatment regimens depend on experimentation. Ciprofloxacin was viewed as advantageous in the treatment of bronchiectasis in a patient with an intriguing condition. Vancomycin is utilized as an experimental treatment for meningitis and extreme fundamental contaminations [57].
Computer Aided Drug Designing (CADD)
Since the 1980s, computer technologies have been used to discover Computer-Aided Drug Designing (CADD) approaches. In 1981, Fortune magazine published an article headlined "Next Industrial Revolution Designing Drugs by Computer at Merck," which highlighted this technology [58]. CADD approaches are divided into two types based on how they are used and whether or not the target structure is available: ligand-based and structure-based methods. Protein Data Bank (PDB) contains an enormous number of tentatively tackled structures that work as homology formats on the off chance that our design of revenue isn't available [59].
Molecular Docking
To conduct activities, proteins recognize other molecular partners. Many biological processes are influenced by Ligand-protein interactions, which has pharmacological implications. Ligand identifies the protein active site's complementary natural shape and binds to it. Because this model was unable to account for many enzyme features such as noncompetitive inhibition and allosteric regulation, it was abandoned [60]. In 1958, Koshland hypothesized enzymatic modifications and coined the term "induced-fit modal" to describe how a ligand causes conformational changes in a protein, resulting in specific ligand-target interactions.Later investigations, on the other hand, explained that an ensemble of conformation in proteins exists naturally, as explored by an energy landscape, and that ligands are preferentially bound to one of them [61].
Prediction of Secondary structure
The PsiPred indicated the nature of proteins and their secondary structure and the results indicated that the protein of Bla-Oxa was consisting of most of the coils and then Helices. Therefore, AATYPE of the protein indicated the nature of protein with maximum regions of non-polar and hydrophobic in nature amino acids. While, in case of SHV protein, it was also found to having most of the region with coils and helices. Although, the AATYPE of the SHV protein indicated the polar nature of the protein but a lot of cysteine residues were found in SHV. The results obtained by PsiPred are given below in the Figure 1.
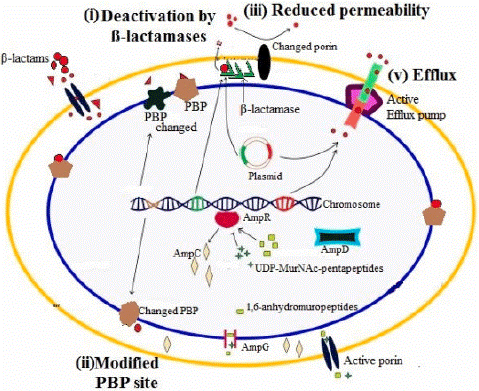
Figure 1: Molecular Mechanism of Β-Lactam Resistance in Gram- Negative bacteria [21].
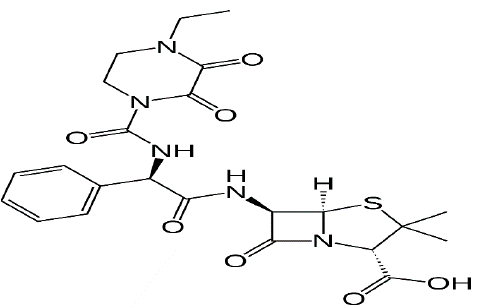
Figure 2: Structural formula of Piperacillin-tazobactam.
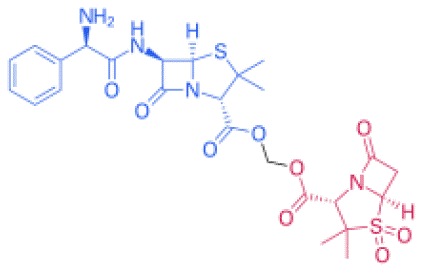
Figure 3: Structural formula of ampicillin sulbactam. Its molecular formula is C8H10NNaO5S.
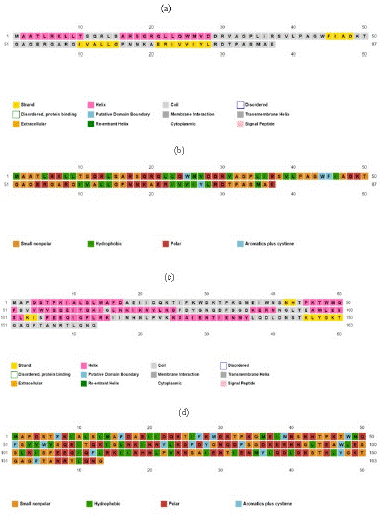
Figure 4: Protein secondary structure analysis by PsiPred.
(a) Secondary structure of Bla-Oxa; (b) Amino Acid Type of Bla-Oxa; (c) Secondary Structure of SHV; (d) Amino Acid Type of SHV
Prediction of Tertiary Structure
The Swiss-Model predicted the tertiary structure of Bla-Oxa with 99.39% sequence identity with a QMEAN Z-Score of -0.69 and GMQE value of 0.97. The scoring index and other relatable factors indicated the protein structure as higher confidence prediction. The quality estimation calculated by Swiss-Model was also in the optimal and standard range. Therefore, the structure was on the quality standards and considerable for the use of docking study and other purposes. The predicted tertiary structure of Bla-Oxa is given below in the Figure 5.
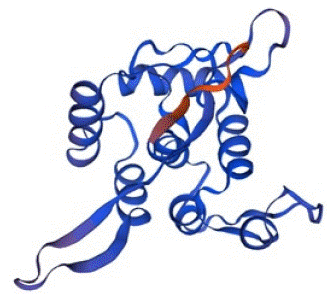
Figure 5: The tertiary structure of Bla-Oxa predicted by Swis-Model with the confidence score of 0.97.
In the same consequences, the structure of SHV was also predicted by Swiss-Model with the percentage identity of 100% and QMEAN Z-score of -0.52. The value of GMQE was calculated as 0.91 with a good confidence score and quality factor. There fore, the protein was also stable and capable to use in various analysis of molecular docking and other related in-silico studies. The predicted structure of SHV is given below in the Figure 6.
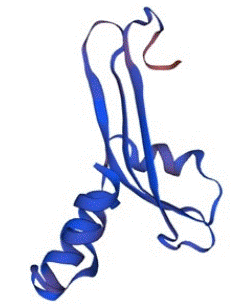
Figure 6: The tertiary structure of SHV predicted by Swiss-Model with the confidence score of 0.91.
Drug Discovery through Molecular Docking
The docking technique by itself has no use, however it can be used in conjunction with experimental and in silico procedures. Several researchers have been upgrading and analysing various docking programs, their performance, and scoring functions after testing has already been completed, resulting in the selection of a certain methodology to hit a specific target system [62].
Docking is used in conjunction with other computational data to obtain information from the P450 system of cytochrome. The bacterial enzyme DNA gyrase performs negative supercoiling and unwinding of bacterial DNA, which is studied as an antibacterial target. HTS was unsuccessful in its quest for possible DNA gyrase protein inhibitors. Using a de novo model, Boehm et al. were able to obtain a significant number of inhibitors for this enzyme [63].
Importance of Computational based Drug Designing
Computer-assisted drug design has made significant progress in predicting biologically active molecules and their receptor binding conformation, with results that are sometimes more exact than those acquired through traditional methods like as high-throughput screening [64].
The results of computational studies are sometimes more precise than the experimental possibilities, and they contribute to the improvement of the experimental array [65].
Discussion
Antibiotic resistance in bacterial pathogen is a great challenge that is linked with high morbidity and mortality. Multi Drug Resistance (MDR) patterns in gram negative and gram positive bacteria are very difficult to treat and may even be untreatable conventional antibiotics. There is currently a shortage of effective therapies, lack of successful precaution measures, and only a few new antibiotics which require development of novel treatment option and alternative antimicrobial therapies. A common cause of antibiotic resistance in bacteria is an increased abundance of Β- lactamases. It can be caused by the selection of resistant variants in the presence of antibiotics.Β- lactamases genes are found in the wild type genomes of many bacteria. There chromosomal Β- lactamases do not generally provide effective antibiotic resistance in wild type bacilli despite evidence that the genes are not completely silenced. Under antibiotic selection pressure however, a number of strains show increased resistance suggesting mutation induced up regulation of Β- lactamases. Microbes can be pathogenic as well as non-pathogenic in nature. They are present everywhere, but our knowledge regarding their diversity is very limited [15].
From history to till date several Strains are enlisted. In this study, molecular , biochemical, phylogenetic and enzyme assay based analysis were used to identify the unknown microbial strains ( three bacterial strains, bacillus cereus, bacillus paramaycoides, Serratia macescens) are their zone of inhibition is measured to check the antibiotic resistance activity [66]. The study conducted for the identification of resistant gene OXA, using pair of primers named as OXA-51 F and OXA-51 R. The product size was 353bp yielded in Acinetobacter bummannii. The gene was also amplified using another pair of primers named as OXA-23 F and OXA-23 R. The product size after sequencing was 501bp in Acinetobacter bummannii [67].
The study conducted in 2019 showed the antibiotic resistance mechanisms in Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterobacter spp. and Acinetobacter spp. The Bla-SHV was amplified using SHV-F and SHV-R primers. The Bla-SHV yielded the amplicon size of 392bp. The Bla-Oxa was amplified using OXA-F and OXA-r primers. The Bla-Oxa gave the amplicon size of 619bp [68].
Conclusion
In the conclusion, beta-lactam is class of antibiotics that is most important and successful drug class however there is no solution that continue to play a valuable role in the fight against the infection caused by this pathogenic bacterium. The molecular analysis and computational analysis of SHV and Bla-OXA is need to show the beta-lactamase resistance and show inhibition against Bacillus and serratia bacterial species. There is need to form a potential compound for the treatment of multi-drug resistant Bacillus and Serratia species infection.
References
- Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of bla (CTX – M), bla (TEM), bla (SHV) and bla (OXA) genes in Extended-spectrum-Β-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrob Resist Infect Control. 2019; 8: 80.
- Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018; 11: 1645-58.
- Chandra S, Prithvi PPR, Srija K, Jauhari S, Grover A. Antimicrobial resistance: call for rational antibiotics practice in India. J Fam Med Prim Care. 2020; 9: 2192-9.
- Bilal H, Khan MN, Rehman T, Hameed MF, Yang X. Antibiotic resistance in Pakistan: a systematic review of past decade. BMC Infect Dis. 2021; 21: 244.
- Gdoura-Ben Amor M, Siala M, Zayani M, Grosset N, Smaoui S, Messadi-Akrout F, et al. Isolation, identification, prevalence, and genetic diversity of Bacillus cereus group bacteria from different foodstuffs in Tunisia. Front Microbiol. 2018; 9: 447.
- Larsson DGJ, Flach CF. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022; 20: 257-69.
- Lobie TA, Roba AA, Booth JA, Kristiansen KI, Aseffa A, Skarstad K, et al. Antimicrobial resistance: A challenge awaiting the post-COVID-19 era. Int J Infect Dis. 2021; 111: 322-5.
- Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Aguilar GR, Gray A. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022; 399: 629-55.
- Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018; 9: 2928.
- Rezasoltani S, Yadegar A, Hatami B, Asadzadeh Aghdaei H, Zali MR. Antimicrobial resistance as a hidden menace lurking behind the COVID-19 outbreak: the global impacts of too much hygiene on AMR. Front Microbiol. 2020; 11: 590683.
- Sawa T, Kooguchi K, Moriyama K. Molecular diversity of extended-spectrum Β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care. 2020; 8: 13.
- Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, et al. Β-lactamases and Β-lactamase Inhibitors in the 21st Century. J Mol Biol. 2019; 431: 3472-500.
- Zeng X, Lin J. Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Front Microbiol. 2013; 4: 128.
- Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010; 54: 969-76.
- Bush K. Past and present perspectives on Β-lactamases. Antimicrob Agents Chemother. 2018; 62: 01076-18.
- Bottone EJ. Bacillus cereus, a volatile human pathogen. Clin Microbiol Rev. 2010; 23: 382-98.
- Fan P, Ma Z, Partow AJ, Kim M, Shoemaker GM, Tan R, et al. A novel combination therapy for multidrug resistant pathogens using chitosan nanoparticles loaded with Β-lactam antibiotics and Β-lactamase inhibitors. Int J Biol Macromol. 2022; 195: 506-14.
- Yamagishi Y, Nakayama N, Matsunaga N, Sakanashi D, Suematsu H, Matsumoto Y, et al. Novel approach for rapid detection of extended spectrum Β-lactamase and metalloid-Β-lactamase using drug susceptibility testing microfluidic device (DSTM). J Infect Chemother. 2022; 28: 526-31.
- Le LTHL, Yoo W, Wang Y, Jeon S, Kim KK, Kim HW, et al. Dual functional roles of a novel bifunctional Β-lactamase/esterase from Lactococcus garvieae. Int J Biol Macromol. 2022; 206: 203-12.
- Hu L, Yang H, Yu T, Chen F, Liu R, Xue S, et al. Stereochemically altered cephalosporins as potent inhibitors of New Delhi metallo-Β-lactamases. Eur J Med Chem. 2022; 232: 114174.
- Ejaz H. Analysis of diverse Β-lactamases presenting high-level resistance in association with OmpK35 and OmpK36 porins in ESBL-producing Klebsiella pneumoniae. Saudi J Biol Sci. 2022; 29: 3440-7.
- Hussain HI, Aqib AI, Seleem MN, Shabbir MA, Hao H, Iqbal Z, et al. Genetic basis of molecular mechanisms in Β-lactam resistant gram-negative bacteria. Microb Pathog. 2021; 158: 105040.
- Zhong D, Zhou Z, Ma W, Ma J, Feng W, Li J, et al. Antibiotic enhances the spread of antibiotic resistance among chlorine-resistant bacteria in drinking water distribution system. Environ Res. 2022; 211: 113045.
- Hutinel M, Larsson DGJ, Flach CF. Antibiotic resistance genes of emerging concern in municipal and hospital wastewater from a major Swedish city. Sci Total Environ. 2022; 812: 151433.
- Huygens J, Rasschaert G, Heyndrickx M, Dewulf J, Van Coillie E, Quataert P, et al. Impact of fertilization with pig or calf slurry on antibiotic residues and resistance genes in the soil. Sci Total Environ. 2022; 822: 153518.
- Onohuean H, Agwu E, Nwodo UU. Systematic review and meta-analysis of environmental Vibrio species–antibiotic resistance. Heliyon. 2022; 8: e08845.
- Bonyadi P, Saleh NT, Dehghani M, Yamini M, Amini K. Prevalence of antibiotic resistance of Pseudomonas aeruginosa in cystic fibrosis infection: A systematic review and meta-analysis. Microb Pathog. 2022; 165: 105461.
- Colautti A, Arnoldi M, Comi G, Iacumin L. Antibiotic resistance and virulence factors in lactobacilli: something to carefully consider. Food Microbiol. 2022; 103: 103934.
- Scaccia N, Vaz-Moreira I, Manaia CM. The risk of transmitting antibiotic resistance through endophytic bacteria. Trends Plant Sci. 2021; 26: 1213-26.
- Ohore OE, Qin Z, Sanganyado E, Wang Y, Jiao X, Liu W, et al. Ecological impact of antibiotics on bioremediation performance of constructed wetlands: microbial and plant dynamics, and potential antibiotic resistance genes hotspots. J Hazard Mater. 2022; 424: 127495.
- Nunziata L, Brasca M, Morandi S, Silvetti T. Antibiotic resistance in wild and commercial non-enterococcal lactic acid Bacteria and bifidobacteria strains of dairy origin: an update. Food Microbiol. 2022; 104: 103999.
- Moser AI, Campos-Madueno EI, Sendi P, Perreten V, Keller PM, Ramette A, et al. Repatriation of a patient with COVID-19 contributed to the importation of an emerging carbapenemase producer. J Glob Antimicrob Resist. 2021a; 27: 267-72.
- Girlich D, Oueslati S, Bernabeu S, Langlois I, Begasse C, Arangia N, et al. Evaluation of the BD MAX Check-Points CPO assay for the detection of carbapenemase producers directly from rectal swabs. J Mol Diagn. 2020; 22: 294-300.
- Madkour LA, Soliman MS, Hassan DM, Soliman NS, ElMahdy YA. Detection of carbapenemase-producers: evaluating the performance of the carbapenem inactivation method and Carba NP test versus multiplex PCR. J Glob Antimicrob Resist. 2017; 9: 10-4.
- Antinori E, Unali I, Bertoncelli A, Mazzariol A. Klebsiella pneumoniae carbapenemase (KPC) producer resistant to ceftazidime–avibactam due to a deletion in the blaKPC3 gene. Clin Microbiol Infect. 2020; 26: 946.e1-3.
- Molnár S, Flonta MMM, Almas A, Buzea M, Licker M, Rus M, et al. Dissemination of NDM-1 carbapenemase-producer Providencia stuartii strains in Romanian hospitals: a multicentre study. J Hosp Infect. 2019; 103: 165-9.
- Bernabeu S, Poirel L, Nordmann P. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn Microbiol Infect Dis. 2012; 74: 88-90.
- Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014; 20: 821-30.
- Zhang H, Liang B, Wang J, Cai Y. Non-carbapenem Β-lactam/Β-lactamase inhibitors versus carbapenems for urinary tract infections caused by extended-spectrum Β-lactamase-producing Enterobacteriaceae: A systematic review. Int J Antimicrob Agents. 2021; 58: 106410.
- Goyal T, Dhamija P, Vardhan G, Gupta P, Trikha V. In vitro study of elution kinetics and biological activity of piperacillin/tazobactam and gentamicin in acrylic bone cement. Orthop Traumatol Surg Res. 2022; 108: 103230.
- Ramassamy M, Murris M, Recoche I, Mailhol C, Didier A, Guilleminault L. Piperacillin-tazobactam-induced fever in chronic respiratory diseases: safe challenge test under premedication. Rev Fr Allergol. 2022; 62: 478-81.
- Nalbant D, Reeder JA, Li P, O’Sullivan CT, Rogers WK, An G. Development and validation of a simple and sensitive LC-MS/MS method for quantification of ampicillin and sulbactam in human plasma and its application to a clinical pharmacokinetic study. J Pharm Biomed Anal. 2021; 196: 113899.
- Uddin TM, Chakraborty AJ, Khusro A, Zidan BRM, Mitra S, Emran TB, et al. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies and future prospects. J Infect Public Health. 2021; 14: 1750-66.
- Lu XM, Chen YL. Varying characteristics and driving mechanisms of antibiotic resistance genes in farmland soil amended with high-density polyethylene microplastics. J Hazard Mater. 2022; 428: 128196.
- Lai CKC, Ng RWY, Leung SSY, Hui M, Ip M. Overcoming the rising incidence and evolving mechanisms of antibiotic resistance by novel drug delivery approaches–an overview. Adv Drug Deliv Rev. 2022; 181: 114078.
- Jiang Q, Feng M, Ye C, Yu X. Effects and relevant mechanisms of non-antibiotic factors on the horizontal transfer of antibiotic resistance genes in water environments: a review. Sci Total Environ. 2022; 806: 150568.
- Song R, Li H, Kang Z, Zhong R, Wang Y, Zhang Y, et al. Surface plasma induced elimination of antibiotic-resistant Escherichia coli and resistance genes: antibiotic resistance, horizontal gene transfer, and mechanisms. Sep Purif Technol. 2021; 275: 119185.
- Li H, Kang Z, Jiang E, Song R, Zhang Y, Qu G, et al. Plasma induced efficient removal of antibiotic-resistant Escherichia coli and antibiotic resistance genes, and inhibition of gene transfer by conjugation. J Hazard Mater. 2021; 419: 126465.
- Subirats J, Murray R, Yin X, Zhang T, Topp E. Impact of chicken litter pre-application treatment on the abundance, field persistence, and transfer of antibiotic resistant bacteria and antibiotic resistance genes to vegetables. Sci Total Environ. 2021; 801: 149718.
- Zhou H, Zhang J, Shao Y, Wang J, Xu W, Liu Y, et al. Development of a high resolution melting method based on a novel molecular target for discrimination between Bacillus cereus and Bacillus thuringiensis. Food Res Int. 2022; 151: 110845.
- Lin Y, Briandet R, Kovács ÁT. Bacillus cereus sensu lato biofilm formation and its ecological importance. Biofilm. 2022; 4: 100070.
- Yu D, Wang N, Gong Y, Wu Z, Wang W, Wang L, et al. Screening of active sites and study on immobilization of Bacillus cereus phospholipase C. LWT. 2022; 159: 113245.
- Le Marc Y, Postollec F, Huchet V, Ellouze M. Modelling the thermal inactivation of spores from different phylogenetic groups of Bacillus cereus. Int J Food Microbiol. 2022; 368: 109607.
- Antil S, Kumar R, Pathak DV, Kumar A, Panwar A, Kumari A. Plant growth-promoting rhizobacteria-Bacillus cereus KMT-5 and B. megaterium KMT-8 effectively suppressed meloidogyne javanica infection. Appl Soil Ecol. 2022; 174: 104419.
- Kaur M, Harpaz D, Eltzov E. Development of a portable colorimetric point-of-care device for the detection of Bacillus cereus in food specimens. Sens Actuators B. 2022; 356: 131354.
- Zheng H, Sheng R, Li H, Ahmad W, Chen Q. Rapid and selective detection of Bacillus cereus in food using cDNA-based up-conversion fluorescence spectrum copy and aptamer modified magnetic separation. Spectrochim Acta A Mol Biomol Spectrosc. 2022; 267: 120618.
- Butcher M, Puiu D, Romagnoli M, Carroll KC, Salzberg SL, Nauen DW. Rapidly fatal infection with Bacillus cereus/thuringiensis: genome assembly of the responsible pathogen and consideration of possibly contributing toxins. Diagn Microbiol Infect Dis. 2021; 101: 115534.
- Ozdemir S, Turkan Z, Kilinc E, Bayat R, Soylak M, Sen F. Preconcentrations of Cu (II) and Mn (II) by magnetic solid-phase extraction on Bacillus cereus loaded γ-Fe2O3 nanomaterials. Environ Res. 2022; 209: 112766.
- Gordon D, Gordon R. CADD: a seamless solution to the domain decomposition problem of subdomain boundaries and cross-points. Wave Motion. 2020; 98: 102649.
- Salmanli M, Tatar Yilmaz GT, Tuzuner T. Investigation of the antimicrobial activities of various antimicrobial agents on Streptococcus mutans sortase A through computer-aided drug design (CADD) approaches. Comput Methods Programs Biomed. 2021; 212: 106454.
- Kang BE, Gall B, Choo E, Sanapareddy N, Rakova I, Keen-Kim JD. Retrospective evaluation of in silico prediction tools, REVEL and CADD, for supporting level evidence (PP3/BP4) of genomic variant interpretation. Mol Genet Metab. 2021; 132: S260-1.
- Kijima K, Mita H, Kawakami M, Amada K. Role of CadC and CadD in the 2, 4-dichlorophenoxyacetic acid oxygenase system of Sphingomonas agrestis 58-1. J Biosci Bioeng. 2018; 125: 649-53.
- Onawole AT, Sulaiman KO, Kolapo TU, Akinde FO, Adegoke RO. COVID-19: CADD to the rescue. Virus Res. 2020; 285: 198022.
- Mather CA, Mooney SD, Salipante SJ, Scroggins S, Wu D, Pritchard CC, et al. CADD score has limited clinical validity for the identification of pathogenic variants in noncoding regions in a hereditary cancer panel. Genet Med. 2016; 18: 1269-75.
- Zhao L, Ciallella HL, Aleksunes LM, Zhu H. Advancing computer-aided drug discovery (CADD) by big data and data-driven machine learning modeling. Drug Discov Today. 2020; 25: 1624-38.
- Patel S, Navas M, Batt C, Jump RL. Oral cryptococcosis in a patient with chronic lymphocytic leukemia. Int J Infect Dis. 2016; 50: 18-20.
- Fenselau C, Havey C, Teerakulkittipong N, Swatkoski S, Laine O, Edwards N. Identification of Β-lactamase in antibiotic-resistant Bacillus cereus spores. Appl Environ Microbiol. 2008; 74: 904-6.
- Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the bla OXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006; 44: 2974-6.
- Abrar S, Ain NU, Liaqat H, Hussain S, Rasheed F, Riaz S. Distribution of bla CTX- M, bla TEM, bla SHV and bla OXA genes in Extended-spectrum-Β-lactamase-producing Clinical isolates: A three-year multi-center study from Lahore, Pakistan. Antimicrob Resist Infect Control. 2019; 8: 80.