
Research Article
Austin J Biotechnol Bioeng. 2023; 10(2): 1126.
Genetic Characterization and Allele Frequencies of Three Populations of Systomus sarana (Hamilton, 1822) of Madhya Pradesh Reveled by Random Amplified Polymorphic DNA
Garg RK¹*; Shweta Mishra²
1Department of Biosciences, Faculty of Life Sciences, Barkatullah University, Bhopal-462026 (M.P.) India
2Awadesh Pratap Singh (APS) University, Rewa (M.P.), India
*Corresponding author: RK Garg Department of Biosciences, Faculty of Life Sciences, Barkatullah University, Bhopal-462026 (M.P.) India. Email: drrkgarg136@gmail.com
Received: October 31, 2023 Accepted: December 04, 2023 Published: December 11, 2023
Abstract
The random amplified polymorphic DNA (RAPD-PCR) technique was examined as a potential tool for identification of genetic stock, gene diversity/gene frequencies studies. During this investigation, total 17 specimens from three study sites i.e., Upper Lake (Bhopal), Kshipra dam (Ujjain) and Wainganga river (Balaghat) were studied and delineated genetic studies and allele frequencies studies among the three populations. Total numbers of multiple loci/DNA fingerprints were 27 in all three populations of which number of polymorphic loci were in 8 in Upper Lake, 11 Kshipra Dam, 8 Wainganga River. Genetic differentiation (GST) among the populations was found to be as 0.5926, estimated gene flow between population as Nm=0.3437, intra-population heterozygosity as HS=0.1518 and total heterozygosity as HT=0.3726 clearly reflecting high genetic polymorphism. Nei’s unbiased genetic identity and genetic distance was obtained as highest distance between Upper Lake and Kishpra dam as 0.7223 however, less genetic distance was obtained between Wainganga River and Khipra dam populations. However, population wise, the genetic analyses in Upper Lake Bhopal indicated lower genetic polymorphism (P) as 29.63% as compared to rest of population’s i.e., Kshipra Dam 40.74% and Wainganga River 62.96%. Nei’s gene diversity (h) was observed as 0.1306 in Upper Lake Bhopal, 0.1288 in Kshipra Dam and 0.1956 Wainganga River reflecting much higher gene diversity in Wainganga River. This may be due to declined of genetic variation in the stocks which could cause by manipulation of habitat as the natural habitat or pressure of anthropogenic activities around the reservoir which may cause the threats of fish habitats.
Keywords: DNA fingerprinting; Gene diversity; Gene frequency; Allele frequencies; Heterogeneity; Haplotype diversity
Introduction
Loss of biodiversity is one of the biggest challenges facing modern society because environmental calamity is increasingly evidenced by the loss or deterioration of genetic resources and habitats, as well as recent attempts to highlight and address the issue at the highest international levels. Despite the enriched biodiversity is accessible species-specific information to assist in sustainable resource management is scarce. Study on genetic diversity with a reliable method to serve the conservation management plan for fish diversity Madhya Pradesh.
Systomus (Puntius) sarana is a tropical freshwater fish belonging to the Puntius genus of cyprinidae family. This species is commonly called as 'olive barb' which can be used both as food fish and ornamental fish. The generic status of the fish is still unclear and keeps flipping between Barbodes and Systomus sarana is a widespread species with no known major widespread threats. Currently, based on its wide distribution and apparent lack of threats it is assessed as Least Concern. However, the species needs to be thoroughly studied before a re-evaluation is done.
Random amplified polymorphic DNA (RAPD-PCR) is a widely used molecular tool in detection and characterization of genetic polymorphisms in natural populations with higher speed and efficiency. RAPD-PCR amplifies DNA segments of variable lengths and such length polymorphisms are inherited in a Mendelian fashion and thus can be used as genetic markers [1]. The main advantage of RAPDs is that they are quick and easy to assay. Because PCR is involved, only low quantities of template DNA are required. Since random primers are commercially available, no sequence data for primer construction are needed. Moreover, RAPDs have a very high genomic abundance and are randomly distributed throughout the genome.
In many studies, random amplified polymorphic DNA (RAPD-PCR) approach served as a powerful and reliable tool for the identification of diversity, comprising all major taxa including marine as well as freshwater species from different geographic regions [2,3].
The present study explored the utility of the RAPD-PCR approaches as a molecular technique for the identification of genetic characterization in Systomus sarana of three population of Madhya Pradesh and evaluated the gene flows success rates based on allele frequencies.
Materials and Methods
Survey and Samples Collection
A detailed survey has been carried out in different three study sites of the major water bodies of the Madhya Pradesh i.e., Upper Lake (Bhopal), Kshipra Dame (Ujjain) and Wainganga River (Balaghat). A total 17 samples of Systomus sarana were collected mainly in summer season (Table 1) with the help of local fishermen and geographic coordinates of the collection sites were recorded as shown in Table 1. The fish specimens were identified with the help of keys [4].
S. No.
Sampling site
Sample code
Geographical location
Sample size
1.
Upper Lake Bhopal
PSBPLACC
23.2532° N, 77.3382° E
n=03
2.
Kshipra Dam Ujjain
PSINDACC
22.9229° N, 75.9774° E
n=07
3.
Wainganga River Balaghat
PSBLACC
19.3524°N, 79.4759°E
n=07
Table 1: Samples information’s of Systomus sarana used for DNA fingerprinting along with their coordinates.
Molecular Study
Collected all the samples were brought the laboratory and dissected, tissue samples were collected for genomic DNA extraction using protocol with slight modification [5]. Then, isolated DNA was introduced to Polymerase Chain Reaction (PCR) using arbitrary random primers. After which, electrophoresis of PCR product has been done, then, obtained the random amplified polymorphic DNA bands on the gel.
The yield of extracted DNA in ng/μl from fish tissues was measured using a UV Spectrophotometer (ND-1000) at 260nm wavelength. The purity of DNA was determined by calculating the ratio of absorbance at 260 nm to that of 280 nm. The ratio of absorption at 260nm v/s 280nm is commonly used to assess the purity of DNA with respect to protein contamination, since protein (in particular, the aromatic amino acid) tends to absorb at 280nm. The DNA sample is considered as pure when the 260 to 280 ratio comes near 1.8. But the DNA sample having ratio 1.5 to 2.0 can be easily used for PCR.
Polymerase Chain Reactions and Agarose Gel Electrophoresis
Ten RAPD (Genei, Banglore, India) primers were primarily scored in 3 individuals from each of the three populations of Puntius sarana. Three primers, which gave polymorphism, were selected for Polymerase Chain Reaction (PCR) and used in the final RAPD analysis of 03individuals collected from Upper Lake (Bhopal),07 Wainganga River (Balaghat) and 07 individuals from Kshipra dam. Those 3 primers namely RAn-1 (AM765834), RAn-2 (AM750059), RAn-3 (AM750052) were used in the present study for RAPD-PCR amplification. The master mix for the PCR was made very carefully and loaded on the thermal cycler for DNA amplification (Table 2). There are three major steps in a PCR, which are repeated for 25 to 40 cycles. This is done on an automated thermal cycler, which can heat and cool the tubes with the reaction mixture in a very short time (Table 3). Obtained PCR product was then subjected to Agarose gel electrophoresis for generating the RAPD bands.
S.No.
Reagent
Amount
Red dye (2x concentration)
15.00 μl
RAPD Primer (10 picomol)
1.20 μl
Template DNA (40-60 ng/μl)
0.60 μl
Sterile Water (Molecular Grade)
13.20 μl
Total volume in μl
30.00
Table 2: Composition of PCR reaction cocktail for amplification of random sites for allele frequency studies.
Initial Denaturation
Step 1
Step 2
Final Extension
Denaturation
Annealing
Extension
Denaturation
Annealing
Extension
94°C
94°C
35°C
72°C
94°C
37°C
72°C
72C
5.0 min.
45 sec.
1.0 min.
1.5 min.
45 sec.
45 sec.
1.0 min.
10.0 min.
Denaturation
X10Cycles
X40 Cycles
Final extension
Table 3: PCR cycles for amplification of random amplicons for allele frequencies studies in Systomus sarana.
Statistical and Bioinformatic Analyses
Reproducibility of RAPD-PCR was tested by gel documentation system. The bands were visualized under UV-light and the gel was photographed. The DNA profile or fingerprint of each fish sample was recorded. The data was scored for the presence or absence of the amplified fragment for all individuals. The data matrix was generated and each individual profile constructed using the following criterions: if a given amplified fragment was present in an individual, then it was assigned as ‘1’ and when the fragment was absent it was assigned as ‘0’. These generated data called as binary matrix which was used for phylogenetic relationship between three populations of Puntius sarana using Past Software (1.91).
Similarity Indexes (SI) were calculated based on the Rf values for individual primers. SI of bands, which were common between two variants, was estimated following Nei and Li (1979). Using Dice coefficient, a similarity matrix involving 6 variants was generated with PAST 1.91 ver software. RAPD is a dominant marker; we assumed that each band represented the phenotype at a single biallelic locus. Therefore amplified fragment were scored for the presence (1) and absence (0) of homologues bands.
A dendrogram is a tree for visual classification of similarity, commonly used in Biology for grouping species. Genetic distance and NMDS was estimated by PAST ver. 1.91. Once we obtained the distance matrix this matrix was used to identify the phylogenetic profiling of three populations using Mega ver 6.0. The allele frequencies and gene diversity was estimated by Popegene ver 3.20 using DNA fingerprinting data as binary matrixes.
Results and Discussion
Population genetic studies are to characterize the extent of genetic variation among and within species and account for this variation which provides useful information on the level of interaction between local populations and permit assessment of the contribution of a meta-population structure to regional persistence of a species [6,7]. The amount of genetic variation within and between populations can be determined by the frequency of genes and the forces that affect their frequencies, such as migration, mutation, selection and genetic drift [8]. During the last two decades, a large amount of genotype and allele frequency data have been obtained from a large number of species, including fish species, through the means of proteomic genomic and molecular techniques. These studies have shown that, most species are sub-divided into more or less distinct units that differ genetically from each other [9].
Isolated genomic DNA was qualitatively and quantitatively estimated by UV–Spectrophotometer (ND-1000). DNA yield and quality were checked by spectrophotometer from the absorbance data of the sample DNA at 260 & 280nm and obtained more than 50 ng/μl concentrations for all 17 samples which have been diluted as per requirements.
Genetic Variability Using DNA Fingerprinting
DNA fingerprints analysis based random primers has been used to elucidate genetic diversity and polymorphism among and within three populations of Systomus sarana of Madhya Pradesh. Genetic differentiation (GST) among the populations was found to be as 0.5926, estimated gene flow between population as Nm=0.3437, intra-population heterozygosity as HS=0.1518 and total heterozygosity as HT= 0.3726 clearly reflecting high genetic polymorphism. The unbiased genetic identity and genetic distance was obtained as highest distance between Upper Lake (Bhopal) and Kishpra dam (Ujjain) as 0.7223 [10], however, less genetic distance was obtained between Wainganga River and Khipra dam populations (Table 4). Total numbers of multiple loci/DNA fingerprints were 27 in all three populations of which number of polymorphic loci were in 8 in Upper Lake, 11 Kshipra Dam, 17 Wainganga River Balaghat. However, population wise, the genetic analyses in Upper Lake indicated lower genetic Polymorphism (P) as 29.63% as compared to rest of population’s i.e; Kshipra dam 40.74% and Wainganga River Balaghat 62.96% (Table 5). Nei’s gene diversity (h) was observed as 0.1306 in Upper Lake, 0.1288 in Kshipra dam and 0.1956 Wainganga River reflecting much higher gene diversity in Wainganga River (Table 5). Same studies on the other freshwater fishes were done by many scientists of India. Some of them have studied on the Heteropneustes fossilis of two populations of which he obtained high polymorphism population as 87.93% [11]. However, in present investigation Systomus sarana showed lower polymorphism as 29.63% in Upper Lake.
Sample Id
Upper Lake Bhopal
Kshipra Dam Ujjain
Wainganga River
BalagahtUpper Lake Bhopal
*****
0.4856
0.4918
Kshipra Dam Ujjain
0.7223
*****
0.8568
Wainganga River Balaghat
0.7096
0.1545
*****
Table 4: Nei's genetic identity (above diagonal) and genetic distance (below diagonal).
S. No.
Population genetic values
Upper Lake Bhopal
Kshipra Dam Ujjain
Wainganga River
Balagaht1.
Sample size (n)
03
07
07
2.
Observed number of alleles (na)*
1.2963±0.4653
1.4074±0.5007
1.6296±0.4921
3.
Effective number of alleles (ne)*
1.2435±0.4027
1.2158± 0.3357
1.3155±0.3329
4.
Nei's (1973) gene diversity (h)*
0.1306±0.2101
0.1288±0.1848
0.1959±0.1832
5.
Shannon's Information index (I)*
0.1867±0.2972
0.1964±0.2673
0.3025±0.2651
6.
Total number of loci
27
27
27
7.
Number of polymorphic loci
8
11
17
8.
% of polymorphic loci
29.63
40.74
62.96 %
Note: mean values with standard deviation (SD)
Table 5: Genetic analysis and allele frequencies among three populations of Systomus sarana Single Population Level (SPL).
In the previous investigation on three species of Garra carried out and obtained high polymorphism [12], if our studies compared with these studies carried; it can be concluded that Systomus sarana having 62.96% polymorphism in Wainganga River Balaghat which is drastically varied in comparison to the other. The analysis of dendrogram was done using software popgene. The tree was constructed by Jaccard’s and Euclidean Paired group method (Figure 4). Dendrogram generated by UPGMA of the fingerprinting indices grouped 17 individuals from the three populations under three clusters which were in agreement with the each population (Figure 4).
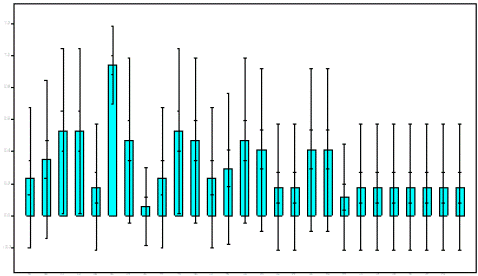
Figure 1: Genetic variations and allele frequencies estimation with standard deviation for 27 alleles of Systomus sarana using DNA fingerprinting.
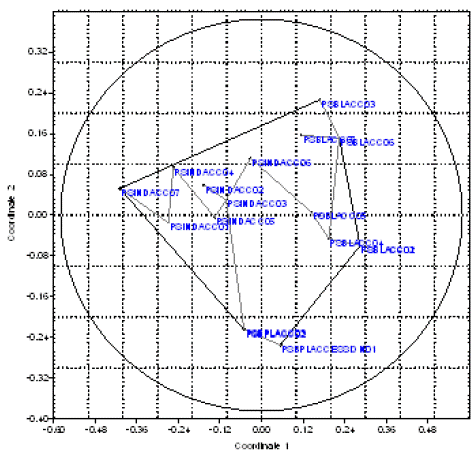
Figure 2: Non-Metric Multi-Dimensional (NMDS) Plot showing scattered genetic variations within three populations using Jaccard’s coefficients.
Figure 3: Genetic frequencies 27 accession of Systomus sarana using DNA fingerprinting as one way ANOVA

Figure 4: Molecular phylogenetic analysis by phylogenetic linkage method as neighbor-joining using DNA fingerprints.
High genetic variation was considered to be related to the adaptive fitness in changing environments, losing genetic variation is detrimental to the domestication process of cultured stocks [13]. We found significantly reduced genetic diversity in terms of Nei’s gene diversity (h) and Shannon Information Index (I) observed in Upper Lake populations (29.63%) when compared to the However, population wise, the genetic analyses in Upper Lake indicated lower genetic Polymorphism (P) as 29.63% as compared to rest of population’s i.e; Kshipra dam and Wainganga River. This may be due to declined of genetic variation in the stocks which could cause by manipulation of habitat as the natural habitat or pressure of anthropogenic activities around the reservoir which may cause the threats of fish habitats.
DNA fingerprinting showed that, compared to Upper Lake, Kshipra and Wainganga River populations, a genetic changes reduced genetic diversity and significant differentiation have taken place in Upper Lake group stocks, as shown by allele richness and heterozygosity studies as well as pairwise Genetic differentiation among the Upper Lake strains could reflect genetic drift due to intensive breeding practices. Thus, in the interests of optimal resource management, genetic variation should be monitored and in-breeding controlled within stocks in commercial breeding programs. Information on genetic population structure based on cross-species markers can aid in the proper management of Systomus sarana populations.
Polymorphism Studies
RAPD markers have been found to have a wide range of application in gene mapping, population genetics, and molecular evolutionary genetics in plant and animal breeding system [14,15]. In Upper Lake, Kshipra dam and Wainganga River, total 27 alleles were obtained in all 3 population of Systomus sarana representing whole Madhya Pradesh for estimation of gene flows and heterogeneity among the populations.
The data for further analyses in the construction of the presence /absence matrix has been constituted. Among the 27 bands that were scored in Wainganaga River Balaghat population 17 (62.96%) were polymorphi loci. Jaccard’s dissimilarity coefficient in 34 individuals of Tenualosa ilisha and observed 20.41% polymorphism [16] and genetic variations of wild and hatchery populations of C. catla revealed by RAPD markers and found overall 54.55% polymorphism [17]. Similarly, we found 84.54% of polymorphic loci in Gandhisagar reservoir which higher than C. catla (75%) [18]. Islam et al. (2005) and in Oreochromis niloticus (55.76%) [19]. However, studies on genetic polymorphism in Clarias batrachus and obtained 72 scorable loci of which 68 (86.66%) were polymorphic showed a highest and significant differences in their degree of polymorphism as compared to our studies [20].
Nei`s genetic identity between Upper lake, Kshipra Dam and Wainganga river was highest between Kshipra Dam and Wainganga river as 0.8568 however was lowest as 0.4856 between Upper Lake and Kshipra dam (Table 7). Genetic diversity as observed no of alleles (na) was 1.2963. Effective number of alleles (ne) 1.2435 and Shannon`s information Index (I) 0.1867.
The protocol for RAPD technique may serve as a reliable, simple, easy to handle and elegant tool in molecular diagnosis of different accessions/variants of an important animal species. In essence, the RAPD method used in this study displayed appreciable intra–population variations (Hs) as 0.2665±0.0217 (Table 4) or molecular polymorphism, which pre-existed in different locations. RAPD analysis has some limitations that must be considered as it has dominant inheritance and marker/marker homozygous cannot be distinguished from marker/null heterozygotes [21]. Further analysis using co-dominant molecular markers like mitochondrial and microsatellite markers is recommended that will further enhance the understanding of the genetic stock structure of Systomus sarana.
On the basis of findings of the present study, it can be concluded that the DNA samples isolated from muscles of Systomus sarana, were of high purity and amenable to further processing in cloning experiments as well as DNA fingerprinting. This optimized protocol for RAPD technique may serve as a reliable, simple, easy to handle and elegant tool in molecular diagnosis of different accessions/variants of an important animal species. In essence, the RAPD method used in this study displayed appreciable inter-population variations or molecular polymorphism, which pre-existed in different locations. In spite of their morphological identity, substantial polymorphism was observed among the variants using RAPD.
Polymorphism assessments were done in 17 individuals of Systomus sarana, 3 individuals collected from Upper Lake and 7 from each Kshipra dam and Wainganga of which polymorphic bands were 27 (64.49%) and 27 bands are generated in populations of Kshipra Dam of which 19 (82.29%) bands are polymorphic. The present study may also serve as a reference point for further examinations of the genetic variation within the population of P. ticto. Furthermore it can be used as a model for other studies relating to genetic diversity. Once their population structure is understood well, the management of this valuable resource could be used to optimize the harvest, as to protect the populations.
The DNA fingerprints for further analyses were scored as presence/absence matrix. Among the 27 bands that were scored in all three populations of which Upper lake having 29.63% with the 8 polymorphic loci. The data for further analyses in the construction of the presence /absence matrix has been constituted. Among the 27 bands that were scored in population of Kshipra dam with 11 (40.74%) were polymorphic loci.
Conclusions
On the basis of findings of the present study, it can be concluded that the DNA samples isolated from muscles of Systomus sarana, were of high purity and amenable to further processing in cloning experiments as well as DNA fingerprinting. This optimized protocol for RAPD technique may serve as a reliable, simple, easy to handle and elegant tool in molecular diagnosis of different accessions/variants of an important animal species. In essence, the RAPD method used in this study displayed appreciable inter-population variations or molecular polymorphism, which pre-existed in different locations. In spite of their morphological identity, substantial polymorphism was observed among the variants using RAPD.
In present study, the variation detected in three populations of Systomus sarana could help in formulating more effective strategies for managing this aquaculture species and also in evaluating the potential genetic effects induced by hatchery operations for selective breeding. The objective of this study is also to assess genetic variations and relatedness in population of Upper Lake, Kshipra dam and Wainganga using RAPD markers. Polymorphism assessments were done in 17 individuals of rana, 3 individuals collected from Upper Lake and 7 from each Kshipra dam and Wainganga of which polymorphic bands were 27 (64.49%) and 27 bands are generated in populations of Kshipra Dam of which 19 (82.29%) bands are polymorphic. The present study may also serve as a reference point for further examinations of the genetic variation within the population of P. ticto. Furthermore it can be used as a model for other studies relating to genetic diversity. Once their population structure is understood well, the management of this valuable resource could be used to optimize the harvest, as to protect the populations.
Author Statements
Acknowledgments
The Authors with thanks of Director General, M.P. Council of Science and Technology, Government of Madhya Pradesh for their help and encouragements during the research work. We wish to thank to local fishermen for helping us during collection of fish samples at sampling sites. The Authors are also with thanking to laboratory colleagues and students for their support during the laboratory support.
References
- Hadrys H, Balick M, Schierwater B. Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Mol Ecol. 1992; 1: 55-63.
- Mark GA, Miller DD, Egan A, Fox J, Greenfield A, Mariani S. DNA barcoding unveils skate (Chondrichthyes: Rajidae) species diversity in ’ray’ products sold across Ireland and the UK. PeerJ. 2013; 1: 1-12.
- Shen Y, Guan L, Wang D, Gan X. DNA bar coding and evaluation of genetic diversity inCyprinidae fish in the midstream of the Yangtze River. Ecol Evol. 2016; 6: 2702-13.
- Shrivastava G 2004. Fishes of UP and Bihar. VishwavidyalayaPrakashan, Chowk. Varanasi. 10th ed. ISBN: 9788171240135, 8171240135.
- Janarthan S, Vincent S. Practical biotechnology, methods and protocols. Vol. 13. India: University Press; 2007; 978-81-7371-582-2.
- Hanski I, Gilpin M. Metapopulation dynamics: brief history and conceptual domain. Biol J Linn Soc. 1991; 42: 3-16.
- Weir BS. Genetic data. Sunderland, MA: Sinauer; Analysis II. 1996.
- Gall GAE. Replicated selection for 21-day pupa weight of Tribolium castaneum. Theor Appl Genet. 1971; 41: 164-73.
- Chakraborty R, Leimar O. Genetic variation within a subdivided population. In: Ryman N, Utter FM, editors. Population genetics and fishery management. Washington: University of Washington. 1987; 89-120.
- Nei M, Li W. Mathematical model for study the genetic variation in terms of restriction endonuclueases. Proc Natl Acad Sci USA. 1979; 74: 5267-73.
- Garg RK, Sairkar P, Silawat N, Vijay N, Batav N, Mehrotra NN. Genetic diversity between two populations of Heteropneustes fossilis (Bloch) using RAPD profile. Int J Zool Res. 2009; 5: 171-7.
- Dhinakaran A, Alikunhi NM, Chinnathambi S, Sornam R, Kalaiselvam M, Rajasekaran R, et al. Assessment of morphometric and genetic variation in three freshwater fish species of the genus Garra(Osteichthyes: Cyprinidae). Not Sci Biol. 2011; 3: 12-6.
- Marja-Liisa K, Tähtinen J, Saisa M, Koskiniemi J. Maintenance of genetic diversity of Atlantic salmon (Salmo salar) by captive breeding programmes and the geographic distribution of microsatellite variation. Aquaculture. 2002; 212: 69-92.
- Bardakci. Random amplified polymorphic DNA (RAPD) markers. Turk J Biol. 2001; 25: 185-96.
- Ertas HB, Seker E. Isolation of Listeria monocystogenes from fish intestines and RAPD analysis. Turk J Vet Anim Sci. 2005; 29: 1007-11.
- Shifat R, Anwara B, Khan H. Use of RAPD fingerprinting for discrimination of two populations of Hilsa shad (Tenualosailisha) from Inland Rivers of Bangladesh.Biochem. Mol Biol. 2003; 36: 462-7.
- Rahman SMZ, Khan M, Islam SAS. Genetic variations of wild and hatchery population of catla Indian Major Carps (Catlacatla Hamilton 1822: Cypriniformes) revealed by RAPD markers. Genet Mol Biol. 2009; 32: 197-201.
- Islam MS, Ahmed ASI, Azam MS, Alam MS. Genetic analysis of three river populations of Catlacatla (Hamilton) using random amplified polymorphic DNA markers. Asian Aust J Anim Sci. 2005; 18: 453-7.
- Zaeem SYE, Ahmed MMM. Genetic differentiation between sex reversal and normal of Full-Sib Nile tilapia, Oreochromis niloticus based on DNA fingerprinting. Res J Fish Hydrol. 2006; 1: 1-5.
- Garg RK, Sairkar P, Silawat N, Batav N, Mehrotra NN. Assessment of genetic diversity of Clarias batrachus (Linn.) in three water bodies of Bhopal using RAPD markers. J Environ Biol. 2010; 31: 749-53.
- Hassanien HA. Use of randomly amplified polymorphic DNA (RAPD) analysis to detect genetic variation in sea bass (Dicentrarchus labrax). J Fish Aquat Sci. 2008; 1: 39-46.