
Research Article
Austin J Biotechnol Bioeng. 2024; 11(1): 1127.
Saccharomyces Cerevisiae Evacuated Cells as a Package Model for a Bioactive Milk Protein
Nawal Abd El-Baky*; Neama Mahmoud Fattouh Rezk; Amro A Amara*
Protein Research Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), Alexandria, Egypt
*Corresponding author: Abd El-Baky N & Amara AA Protein Research Department, Genetic Engineering and Biotechnology Research Institute (GEBRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg El-Arab City, PO Box 21934 Alexandria, Egypt. Tel: +2034593422; Fax: +2034593407 Email: nelbaky@srtacity.sci.eg; nawalabdelbaky83@gmail.com; amroamara@web.de, aamara@srtacity.sci.eg
Received: December 29, 2023 Accepted: February 01, 2024 Published: February 07, 2024
Abstract
Protein therapeutics have enlarged susceptibility to degradation/denaturation if compared to small molecules therapeutics, leading their delivery to be demanding. Saccharomyces cerevisiae have the benefits of being superior in size and less antigenic than bacteria, hence was applied in encapsulation or loading of various drugs to treat diseases. Evacuated microbes were employed as carrier for drug delivery. In this study, S. cerevisiae evacuated cells were utilized for packing of camel milk lactoferrin (cLf). Plackett-Burman experimental design was conducted to randomize four variables for cell evacuation. These variables are Sodium Dodecyl Sulfate (SDS, X1), NaOH (X2), NaHCO3 (X3), and H2O2 (X4). The best obtained experiment conditions were used to prepare yeast evacuated cells. Each of cell quality %, and released protein or DNA was estimated. Evacuated cells were then packed with cLf. The packing was validated by scanning electron microscopy. Antifungal efficacy of yeast evacuated cells’ delivery system for cLf against Candida albicans was checked. Its Minimum Inhibitory Concentration (MIC) was calculated. A standard antifungal agent (amphotericin B) was also packed in evacuated yeast. This packed standard was employed as positive control in anti-Candida assay. The MIC for free cLf was 1.3 mg/ml and for free amphotericin B was 5 μg/ml. Those of packed cLf and amphotericin B were 2.6 mg/ml and 10.5 μg/ml, respectively. This work introduced a safe delivery system for cLf that protected the protein and its bioactivity and can be used for other proteins’ applications. Development to topical formulations is also recommended.
Keyword: Anti-Candida; Antifungal; Camel milk lactoferrin; Candida albicans, Drug delivery system; Yeast evacuated cells
Abbreviations: cLf: Camel Milk Lactoferrin; Lf: Lactoferrin; MGC: Minimum Growth Concentration; MIC: Minimum Inhibitory Concentration; SDS: Sodium Dodecyl Sulfate; SDS-PAGE: Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis
Introduction
Lately, manipulating drug delivery systems has considerably evolved because of nanoscience expansion. It has some advantages based on their slower drug release and drug protection nature [1]. For examples; it was used for evading prompt in vivo clearance, unspecific absorption, and destroys biological barriers. Drug delivery systems based on yeast are among the most common systems for targeted delivery [1-3]. Yeast was applied in encapsulation of insoluble drugs of small molecules, nucleic acid, polymers, numerous nanoparticles, and liposomes for disease treatment [4]. For instance, berberine was encapsulated in S. cerevisiae for application in diverse drug and food manufacturing [5]. Fully evacuated microbes (ghosts) possess various applications in Biotechnology. Engineered cells that express particular foreign antigens on their surface were evacuated and then utilized as vaccines [6]. On the other hand, evacuated cells were used as drug delivery systems [7]. There are many approaches for evacuating microbial cells, among them the recent approach developed in the past decade, known as the sponge-like protocol [8,9]. Examples about adopting evacuated cells as advanced drug delivery system include bacterial evacuated cells that were employed as drug carriers directed by cell membrane effect on loading of drugs [10]. Bacterial evacuated cells from Mannheimia haemolytica were involved in delivery of doxorubicin to human cancer cells (Caco-2) in a site-specific way [7]. E. coli and S. cerevisiae evacuated cells were applied as a drug carrier for a therapeutic polyphenolic compound; gossypol acetic acid [11,12].
Bioactive proteins are a vital type of therapeutics. Yet, their delivery is demanding due to their susceptibility to modifications in structure, sensitivity to proteases-mediated hydrolysis, immunogenicity, as well as restricted uptake into cells [13]. To conquer these obstacles, proteins were loaded in several nanoparticles or extracellular vesicles for their delivery. Nevertheless, these delivery approaches have the drawbacks of protein damage and cytotoxicity [14-16].
Lactoferrin (Lf) is a bioactive milk protein [17]. It has a molecular weight of around 80 kDa, and is soluble in water. The parent protein, as well as its-derived bioactive peptides have antibacterial besides antifungal effects [18,19]. Lactoferrin or its-derived peptides either alone or in combination with antifungal agents have anti-Candida efficacy [20,21]. In this work, we employed yeast evacuated cells in packing of camel milk lactoferrin. Plackett-Burman randomization was used to randomize four variables for cell evacuation to guarantee the best yeast evacuated cells preparation. The yeast evacuated cells packed with cLf were tested for their anti-Candida effectiveness.
Material and Methods
Preparation of Evacuated Cells from S. Cerevisiae
Commercial instant dry yeast (S. cerevisiae manufactured by Pakmaya, Turkey) was bought from resident marketplace in Egypt. Plackett-Burman twelve experimental randomization was conducted to spot the best conditions for preparing yeast evacuated cells [22]. The involved four variables in this randomization were the concentrations of four chemical agents, SDS (X1), NaOH (X2), NaHCO3 (X3), and H2O2 (X4). The four variables were randomized as either high (+1/MIC) or low (-1/minimum growth concentration (MGC)) according to Amara (2015) [12]. The cells quality (stated as %) for each experiment was estimated through light microscopy. The total released protein or DNA from evacuated cells was valued spectrophotometrically.
About half gram of the dry yeast was added to 5 ml of either MIC or MGC (according to randomization values) of SDS, NaOH, and NaHCO3, and was shaken gently for 30 min. After centrifugation at 1000 rpm for 5 min, the supernatant was kept, and the concentrations of protein and DNA in this supernatant were quantified at 280 nm and 260 nm, respectively. The cells were washed with sterile double-distilled water, centrifuged at 1000 rpm for 5 min. The same steps were repeated for H2O2. Subsequently, the cells were washed with 60% ethanol, centrifuged at 1000 rpm for 5 min, and the supernatant was tested spectrophotometrically at 280 nm and 260 nm, respectively. Furthermore, the supernatant collected from SDS/NaOH/NaHCO3 treatment step was added to that collected from H2O2 step, then precipitated and washed via cold ethanol (for DNA electrophoresis) or acetone (for protein electrophoresis). The obtained pellet then resolved on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and 2% agarose gel to examine the total released proteins and DNA, respectively, from evacuated cells.
Microscopic Inspection of Yeast Evacuated Cells
Evacuated yeast cells were stained with methylene blue. In short, cells were blended thoroughly with equal volume of 0.1% (w/v) methylene blue solution. The combination was left (5 min) to react, and then cells were inspected by light microscope. Evacuated yeast cells quality was evaluated as % based on count of dead cells to live ones.
Packing the Evacuated Yeast Cells by Camel Milk Llactoferrin
The best attained evacuated cells from the twelve-randomization were used for packing of cLf. Camel lactoferrin was purified from milk as earlier detailed by Redwan and Tabll (2007), and sterilized via 0.22 μm syringe filter [23]. Evacuated yeast cells were added to cLf at concentration of 20 mg/ml, incubated on ice for 2 h with gentle shaking, followed by opening of the tube under sterile air flow for 30 min to dry the cells. Amphotericin B solution at concentration of 25 μg/ml dissolved in ethanol was purchased from EuroClone (Pero, Italy). Evacuated yeast cells were added to 2 ml of amphotericin B solution, incubated at room temperature for 30 min, followed by opening of the tube under sterile air flow for 30 min to let ethanol to evaporate. Drug delivery system of amphotericin B was used as antifungal standard.
Electron Microscope Scanning of Evacuated Yeast Cells Packed with cLf
Previously prepared camel lactoferrin at a concentration of 2 mg/ml in 5 mM sodium carbonate buffer of pH 9.4 and labeled with gold nanoparticles (AuNPs) was utilized in this analysis [24]. Evacuated yeast cells were added to AuNPs-labeled cLf, incubated on ice with gentle shaking for 1 h, dried for 30 min, washed briefly with sterile double-distilled water and spin for 30 seconds, followed by opening of the tube under sterile air flow for 30 min to dry the cells. Cells packed with cLf following each of the above-mentioned steps were studied using a JEOL JSM-IT200 scanning electron microscope. Evacuated yeast cells without any packing were used as control.
Evaluation of Amphotericin B in Evacuated Yeast Cells
Different concentrations from the standard amphotericin B (25 μg/ml in ethanol) varied between 0.31 and 10 μg/ml were set up. The absorbance was recorded at 408 nm [25]. From attained data, the calibration curve for amphotericin B was drawn. The absorbance of amphotericin B packed in evacuated cells after resuspension of dried evacuated yeast carrying the drug in ethanol for 2 h, was estimated at 408 nm, and then its concentration was evaluated from the calibration curve.
Evaluation of cLf in Yeast Evacuated Cells
Dried evacuated cells delivery system for cLf was resuspended in 10 mM PBS at pH of 8.0 for 2 h on ice. Then the cells were removed after centrifugation and the supernatant was kept. The protein in supernatant was quantified by Bradford assay [26].
Broth Microdilution Assay for Anti-Candida Activity of Evacuated Cells Delivery Systems
The used pathogen to evaluate antifungal activity of evacuated cells delivery systems for amphotericin B, or cLf was Candida albicans ATCC 10231 from Becton Dickinson (France). C. albicans was grown in potato dextrose broth at 30 °C for 24 h to refresh the cells.
To assess antifungal activity of evacuated cells delivery systems against C. albicans and calculate their MIC values, broth microdilution technique was employed. Two-fold serial dilutions from evacuated cells delivery system for amphotericin B (10.5, 5.25, 2.62, 1.31, and 0.65 μg/ml in potato dextrose broth) and evacuated cells delivery system for cLf (5.2, 2.6, 1.3, 0.65, and 0.325 mg/ml in potato dextrose broth) were added to inoculated plate with C. albicans. The same dilutions from free (unpacked) amphotericin B, or cLf were used as positive controls. While, yeast evacuated empty cells (without any packing) was the negative control. After incubation at 30°C for 24 h, OD at 600 nm was checked to assess C. albicans growth. Test was performed in triplicates. The lowest concentration that revealed complete inhibition of C. albicans growth was reflected as MIC for each tried antimicrobial.
Results
Analysis of Plackett-Burman Experiments for S. cerevisiae Evacuated Cells
Table 1 indicated the results of released protein and DNA of 12 experiments of Plackett-Burman experimental randomization for each step-in preparation of evacuated cells from S. cerevisiae. The evacuated cells quality (stated as %) and total released DNA and protein of twelve Plackett-Burman experiments for S. cerevisiae evacuated cells preparation in Table 2 were explored by multiple regression analysis using Statgraphics Centurion XV version 15.2.11.
Experiment No.
SDS/NaOH/NaHCO3
H2O2
Ethanol
Protein
DNA
Protein
DNA
Protein
DNA
(mg/ml)
(μg/ml)
(mg/ml)
(μg/ml)
(mg/ml)
(μg/ml)
1
4.44
165.5
1.22
13
0.64
8.75
2
4.58
196.35
1.41
19
0.54
5.95
3
3.15
105.8
1.33
12.75
0.51
5.05
4
4.29
209.35
1.24
14
0.39
4.15
5
4.11
182.5
1.49
38
0.15
6.92
6
4.58
206.8
1.64
14.3
0.21
8.75
7
4.38
201.9
1.28
7.45
0.51
3.65
8
4.21
189.4
3.72
56.5
0.28
11.9
9
3.89
173.1
1.22
9.3
0.31
1.2
10
4.11
178.35
1.17
36.2
0.2
6.07
11
4.09
167.85
1.75
26.85
0.16
0.8
12
4.62
150.55
1.29
25.2
0.28
4.25
Table 1: The results of released protein and DNA of Plackett-Burman randomization for each step-in preparation of evacuated cells from Saccharomyces cerevisiae.
Experiment No.
Variables
Cell quality %
Total DNA (μg/ml)#
Total protein (mg/ml)@
Step no 1$
Step no 2^
SDS
NaOH
NaHCO3
H2O2
(X1)
(X2)
(X3)
(X4)
g/ml
g/ml
g/ml
ml/ml
1
0.01 (+1)
0.01 (-1)
0.1 (+1)
0.3 (+1)
95
189.55
6.36
2
0.001 (-1)
0.01 (-1)
0.01 (-1)
0.15 (-1)
80
213.25
6.3
3
0.01 (+1)
0.1 (+1)
0.1 (+1)
0.15 (-1)
85
126.72
4.54
4
0.001 (-1)
0.1 (+1)
0.01 (-1)
0.15 (-1)
85
256.1
5.99
5
0.001 (-1)
0.1 (+1)
0.1 (+1)
0.3 (+1)
65
200.45
6.26
6*
0.01 (+1)
0.1 (+1)
0.01 (-1)
0.3 (+1)
80
226.15
6.14
7
0.001 (-1)
0.01 (-1)
0.01 (-1)
0.3 (+1)
85
259.6
8.41
8
0.001 (-1)
0.1 (+1)
0.1 (+1)
0.15 (-1)
70
204.77
5.63
9*
0.01 (+1)
0.1 (+1)
0.01 (-1)
0.3 (+1)
78
210.1
5.22
10
0.001 (-1)
0.01 (-1)
0.1 (+1)
0.3 (+1)
85
209.45
6.14
11
0.01 (+1)
0.01 (-1)
0.01 (-1)
0.15 (-1)
85
193.05
5.38
12
0.01 (+1)
0.01 (-1)
0.1 (+1)
0.15 (-1)
65
150.55
4.62
$Step no 1 includes a mixed treatment with SDS, water, NaOH, NaHCO3. Water must be used to avoid chemical reactions.
^Step no 2 includes H2O2 treatment (H2O2 must not be used in step 1).
*Experiments No. 6 and No. 9 (bold) have the same +1 and -1 distribution and were used to confirm the efficiency of the design and obtained results.
#Total DNA (μg/ml) obtained from step 1 and step 2 as well as from their washing step with ethanol.
@Total protein (mg/ml) obtained from step 1 and step 2 as well as from their washing step with ethanol.
Table 2: Evacuated cells quality and total released DNA and protein attained from Plackett-Burman randomization for preparation of evacuated cells from Saccharomyces cerevisiae.
Main effect results of the four variables involved in Plackett-Burman design of evacuated yeast preparation on evacuated cells quality and total released DNA and protein were shown in Figure 1 and Table 3.
Variables
Values
Unit
Main effect ∑(+1)/n(+1)-∑(-1)/n(-1)
(∑+1)/ n(+1)
(∑-1)/ n(-1)
Cell quality %
SDS
488
470
g/ml
1.5
NaOH
463
495
g/ml
-2.66667
NaHCO3
465
493
g/ml
-2.33333
H2O2
488
470
ml/ml
1.5
Total released DNA
SDS
182.6867
223.9367
g/ml
-3.4375
NaOH
204.0483
202.575
g/ml
0.122778
NaHCO3
180.2483
226.375
g/ml
-3.84389
H2O2
215.8833
190.74
ml/ml
2.095278
Total released protein
SDS
5.376667
6.455
g/ml
-0.08986
NaOH
5.63
6.201667
g/ml
-0.04764
NaHCO3
5.591667
6.24
g/ml
-0.05403
H2O2
6.421667
5.41
ml/ml
0.084306
Table 3: Main effect of the four variables involved in Plackett-Burman randomization of evacuated yeast cells preparation on cell quality, and total released DNA and protein.
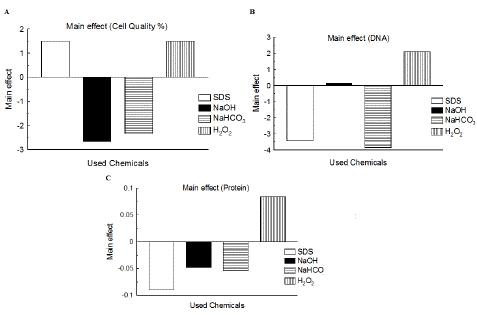
Figure 1: Main effect of the four variables involved in Plackett-Burman randomization of evacuated yeast cells preparation on evacuated cells quality (stated as %) and total released DNA and protein. A: main effect of the four variables on cell quality %; B: main effect of the four variables on total released DNA; C: main effect of the four variables on total released protein.
The created model from Plackett-Burman design via multiple regression was built on the 1st order-model [22]:
In this model, the anticipated response is represented as Y, model constant as Β0, and linear coefficient of variables as Βi. Generated models of multiple regression investigation of 12 experiments of evacuated yeast cells preparation were as follows:
DNA (μg/ml) = 203.312 - 20.625*SDS + 0.736667*NaOH - 23.0633*NaHCO3 + 12.5717*H2O2
Protein (mg/ml) = 5.91583 - 0.539167*SDS - 0.285833*NaOH - 0.324167*NaHCO3 + 0.505833*H2O2
Cell Quality % = 79.8333 + 1.5*SDS - 2.66667*NaOH - 2.33333*NaHCO3 + 1.5*H2O2
The confidence level (%) of gained P-value from multiple regression investigation of data of the four variables (SDS, NaOH, NaHCO3 and H2O2) for each of cell quality, and total released DNA and protein was considered using Statgraphics Centurion XV (Table 4). In case of confidence level % from multiple regression exploration of achieved cell quality %, none of the variables gave greater or equal to 95.0% as showed in Table 4. Regarding confidence level % of total released DNA, three variables had confidence level % >95.0% (SDS of 99.49%, NaHCO3 of 99.71%, and H2O2 of 95.55%) as in Table 4 and thus considered as significant variables. NaOH showed confidence level % equal to 10.98, which is not significant.
Estimate
Standard error
T Statistic
P-Value
Confidence level %
Cell quality%
CONSTANT
79.8333
2.88469
27.6749
0.0000
100
SDS
1.5
2.88469
0.519987
0.6191
38.09
NaOH
-2.66667
2.88469
-0.924421
0.3860
61.4
NaHCO3
-2.33333
2.88469
-0.808868
0.4452
55.48
H2O2
1.5
2.88469
0.519987
0.6191
38.09
Total released DNA
CONSTANT
203.312
5.14482
39.5177
0.0000
100
SDS
-20.625
5.14482
-4.00888
0.0051
99.49
NaOH
0.736667
5.14482
0.143186
0.8902
10.98
NaHCO3
-23.0633
5.14482
-4.48282
0.0029
99.71
H2O2
12.5717
5.14482
2.44356
0.0445
95.55
Total released protein
CONSTANT
5.91583
0.16956
34.8893
0.0000
100
SDS
-0.539167
0.16956
-3.17979
0.0155
98.45
NaOH
-0.285833
0.16956
-1.68573
0.1357
86.43
NaHCO3
-0.324167
0.16956
-1.91181
0.0975
90.25
H2O2
0.505833
0.16956
2.98321
0.0204
97.96
Table 4: Multiple Regression examination of cell quality, and total released DNA and protein against four variables in Plackett-Burman randomization of evacuated yeast cells preparation.
In case of confidence level % of total released protein, two variables had confidence level % >95.0% (SDS of 98.45% and H2O2 of 97.96%) as in Table 4 and consequently considered as significant variables. NaOH and NaHCO3 exhibited confidence level % equal to 86.43 and 90.25, respectively, which are not significant. Yet, in line with Stowe and Mayer (1966), variables with confidence level % of 70%-90% were reflected as being effective [27]. In that case, NaOH (confidence level % equal to 86.43) and NaHCO3 (confidence level % equal to 90.25) could be reflected as effective variables. Since the P-value in the analysis of variance (ANOVA) (Table 5) of cell quality % was 0.7297, which is greater than 0.05, there was not a statistically significant association between the variables at 95.0% or greater confidence level. Regarding total released DNA and protein, the P-values in Table 5 were 0.0044 and 0.0174, respectively, which are less than 0.05, revealing (for each) a statistically significant association between the variables at the confidence level of 95.0%.
Source
Sum of squares
Df
Mean square
F-ratio
P-value
Cell quality %
Model
204.667
4
51.1667
0.51
0.7297
Residual
699.0
7
99.8571
Total (Corr.)
903.667
11
Total released DNA
Model
13390.8
4
3347.69
10.54
0.0044
Residual
2223.41
7
317.631
Total (Corr.)
15614.2
11
Total released protein
Model
8.80023
4
2.20006
6.38
0.0174
Residual
2.41506
7
0.345008
Total (Corr.)
11.2153
11
Table 5: ANOVA for the variables used to optimize cell quality %, and total released DNA and protein.
SDS-PAGE and Agarose Gel Examination of Released Protein and DNA
The supernatant gotten from SDS/NaOH/NaHCO3 treatment step was added to that collected from H2O2 step for each evacuated yeast cells preparation experiment of 12 Plackett-Burman experiments, then resolved on 12% SDS-PAGE (Figure 2). Figure 2 revealed that proteins were released from all evacuated yeast cells preparation experiments, especially experiment No. 1 that showed profile of dense protein bands. Figure 3 displayed 2% agarose gel electrophoresis of released DNA from twelve evacuated yeast cells preparation experiments.

Figure 2: SDS-PAGE of released protein from S. cerevisiae evacuated cells prepared in 12 Plackett-Burman experiments. Lanes M: pre-stained protein marker; Lane C: live S. cerevisiae boiled in sample buffer of SDS-PAGE for 2 min and loaded on gel as control; Lanes 12-1: represent evacuated cells preparation experiments No. 12-1, respectively.
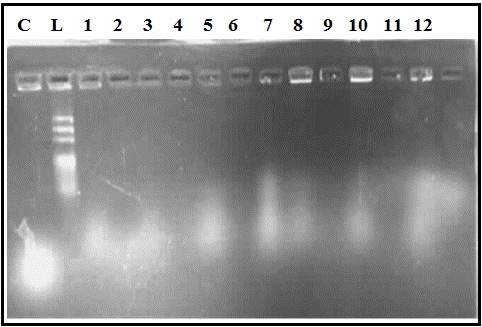
Figure 3: Agarose gel electrophoresis of released DNA from S. cerevisiae evacuated cells prepared in 12 Plackett-Burman experiments. Lane L: DNA ladder (100-1500 bp); Lane C: live S. cerevisiae lysed with DNA extraction buffer as control; Lanes 1-12: represent evacuated cells preparation experiments No. 1-12, respectively.
Light Microscope Investigation of Evacuated Yeast Cells
Examination of S. cerevisiae evacuated cells from twelve Plackett-Burman experiments under light microscope at 40X magnification revealed that evacuated yeast were stained with methylene blue and appeared as dark blue cells approving their death (Figure 4). On the other hand, live S. cerevisiae was not stained since methylene blue can exclusively stain dead cells. Evacuated yeast cells quality was calculated based on count of dead cells to live ones. Accordingly, the best cell quality (95%) was achieved from experiment No. 1 (Table 2).
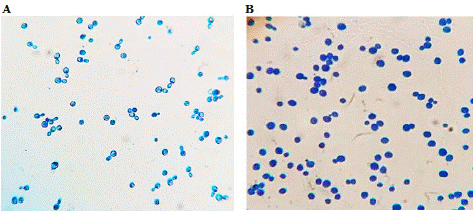
Figure 4: Examination of S. cerevisiae evacuated cells from the best Plackett-Burman experiment under light microscope at 40X magnification. A: live S. cerevisiae as control (did not take the stain); B: evacuated/dead yeast cells from experiment No. 1 stained with methylene blue and appeared as dark blue cells.
Remarkably, experiments No. 6 and 9, which were used as internal control (had the same +1 and -1 distribution to validate the design efficiency and achieved results) in the Plackett-Burman design gave comparable results for each of cell quality %, and total released DNA (Table 2).
Electron Microscope Scanning of Yeast Evacuated Cells Packed with cLf
All of the above-mentioned data suggested that the best attained yeast evacuated cells preparation was from experiment No. 1. As a consequence, yeast evacuated cells from experiment No. 1 were used as delivery system for packing of cLf. Cells packed with cLf were checked via scanning electron microscope in comparison to yeast evacuated empty cells without any packing (Figure 5).
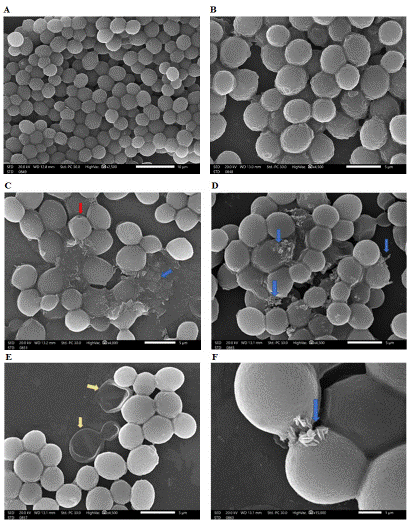
Figure 5: Inspection of evacuated yeast cells delivery system for cLf via scanning electron microscopy. A and B imply control evacuated cells prior to packing of protein; C and D signify evacuated yeast cells during the protein packing process, E signifies successful packing and washing of excess protein (but two yeast evacuated cells were not packed), F represents packed cells with some protein still on their surface (or may be coming out of cells). Blue arrows showed protein crystals, yellow arrows showed empty evacuated cells, red arrow showed pore in evacuated yeast cells.
Figure 5 revealed that yeast evacuated cells had each a tiny small pore, which is accountable for their evacuation. The cells also retained their 3D structure. The images of empty evacuated cells before packing of cLf (Figure 5A and 5B) exhibited that cells had correct 3D structure; meanwhile their surfaces were not smooth where wrinkles appeared on the entire cell surface due to their loss of the cytoplasm. The images of yeast evacuated cells during the packing process following drying and before washing (Figure 5C and 5D) showed protein outside the cells. After packing of the evacuated cells with cLf, their 3D structure was different. Figure 5E and 5F displayed washed and dried S. cerevisiae evacuated cells delivery system for cLf with successful elimination of the protein in the background.
Assessment of Amphotericin B in Evacuated Yeast Cells
Figure 6 illustrated the calibration curve for amphotericin B prepared from different concentrations of the antifungal agent in ethanol and absorbance valuation at 408 nm.

Figure 6: The calibration curve for amphotericin B.
Absorbance of amphotericin B packed in evacuated cells at 408 nm was 1.56±0.05, thus its calculated concentration was 10.5 μg/ml.
Evaluation of cLf in Evacuated Yeast Cells
The supernatant from dried evacuated yeast cells packed with cLf, resuspended in PBS, and centrifuged, was checked for protein concentration through Bradford assay. The achieved cLf concentration packed in evacuated yeast cells was 5.2 mg/ml.
Broth Microdilution Assay for Anti-Candida Efficacy of Evacuated Cells Delivery Systems
MIC against C. albicans for unpacked or free cLf was 1.3 mg/ml, and unpacked or free amphotericin B was 5 μg/ml, while MIC values of packed cLf and amphotericin B were 2.6 mg/ml and 10.5 μg/ml, respectively. This means that MIC values for evacuated cells delivery systems for cLf and amphotericin B increased twice against C. albicans. These findings suggested the decrease in anti-Candida efficacy for protein in evacuated cells delivery system compared to free cLf, which may be due to the slower release rate of cLf from evacuated cells delivery system.
Discussion
Yeasts are viewed historically as the leading microbial cells utilized by human to process foods and make alcoholic beverages. Currently, baker yeast (S. cerevisiae) is more than bread or alcoholic beverages key contributor; it has countless biotechnological as well as pharmaceutical applications. Unicellular microbes including yeast are less immunogenic than other microbes, consequently can be a potential efficient drug delivery system. Moreover, because of their exceptional targeting mechanism specific for phagocytes besides being less immunogenic, drug delivery systems based on yeast may operate via diverse routes of administration [4,28].
Yeast was applied as an efficient drug carrier via its use in encapsulation of functional RNA [29], DNA [30], liposome [31], small molecules drugs [32], specific antigens [33], and others. S. cerevisiae was introduced as a novel carrier for berberine [5]. In contrast to those drugs, bioactive proteins are macromolecules that have unique prerequisite concerning their bioavailability, toxicity, bioactivity, immunogenicity, solubility, precipitation, binding to membranes, and the like. Yeast was used as expression system for the synthesis of recombinant proteins or antigen target proteins, then applied as whole recombinant yeast vaccine [34].
Microbial ghosts are evacuated microbes from their cytoplasm but still keeping their 3D structure. Evacuated microbes were implemented in vaccine preparation for the evacuated microbe itself or used as drug delivery systems [6,7,10]. There are numerous approaches to evacuate microbial cells [8,9,35,36].
In the present study, S. cerevisiae evacuated cells were prepared by an optimized sponge-like procedure to be used as delivery system for a bioactive milk protein of 80 kDa molecular weight; cLf. The packed protein in yeast evacuated cells was verified for anti-Candida activity against C. albicans to confirm that it retains its bioactivity after packing and found to have inhibitory impact on the pathogen.
One of the authors in this work was among the team who developed the sponge-like approach to evacuate microbes [8]. They developed the protocol employing critical concentrations (MIC and MGC) of definite chemicals or proteins (e.g. lysozyme) that permit pore (s) induction in cell wall of microorganisms leading to evacuation of their cells. The protocol was previously effectively applied to develop S. cerevisiae evacuated cells and pack evacuated cells with gossypol acetic acid but the delivery system was not optimized besides incubation time and conditions with evacuation chemicals were different from those implied in this study [12,37].
In the current study, twelve Plackett-Burman experimentations were carried out to explore the effect of four variables on S. cerevisiae evacuated cells quality (stated as %) and total released DNA and protein from yeast evacuated cells. The implicated four variables in this design were SDS (X1), NaOH (X2), NaHCO3 (X3), and H2O2 (X4). One should take into consideration that the effect of each variable on cell quality %, and total released DNA and protein varies with respect to the way each chemical affects microbial cell.
SDS can disturb cell membranes of yeast and prompts the signaling pathway of cell wall integrity [38, 39]. It also can lead to release of cellular proteins from yeast [40]. Sodium hydroxide was used (mixed in a buffer) to prepare yeast cells for extraction of their whole protein content [41]. On the other hand, hydrogen peroxide could inactivate and cause mutation in yeast cells, as well as affecting their protein synthesis [42,43]. NaHCO3 was used in this work rather than CaCO3 in the original sponge-like protocol to evacuate bacterial cells, since it is more potent against eukaryotic cells as demonstrated by Amara et al. (2013) [8] and Amara (2015) [37]. NaHCO3 acts synergistically with SDS to cause damage in yeast cell wall.
Additionally, the concentration of released DNA and protein from step 1 (SDS/NaOH/NaHCO3) in S. cerevisiae evacuation affects that of released DNA and protein from step 2 (H2O2), thus we analyzed experimental data of total released DNA and protein rather than step by step.
With regard to confidence level % calculated in this study from multiple regression exploration of total released DNA from yeast evacuated cells, three variables (SDS (X1), NaHCO3 (X3), and H2O2 (X4)) had confidence level % >95.0% and hence reflected as significant variables. In contrast, data of confidence level % of achieved cell quality % revealed that none of the variables gave greater or equal to 95.0% (i.e. all reflected as non-significant variables). This established our preceding argument that cell wall of microorganisms is more resistant to the implicated chemicals than DNA [8].
Camel lactoferrin was chosen in the present work to be packed in yeast evacuated cells delivery system because of its multiple and diverse bioactivities and superiority in its antimicrobial effectiveness than lactoferrin from other animal species or even human [44]. Furthermore, it has large size (around 80 kDa) and could bind to membrane proteins of bacteria and other microbes [44]. It possesses significant anti-Candida activity against C. albicans either alone or in combination with other antimicrobials such as oleic acid [21]. We found that the protein has retained its bioactivity after packing into evacuated yeast. MIC value for evacuated cells delivery system for cLf increased twice against C. albicans compared to that of free protein, which may be because of the slower release rate of cLf from evacuated cells delivery system.
Scanning electron microscope images of evacuated yeast cells delivery system for cLf in this work revealed some interesting remarks; some evacuated cells did not load the protein and thus appeared transparent and wrinkled or even ruptured because of their emptiness, packed evacuated cells have more correct 3D structure, the protein outside cells in the background significantly disappeared after washing, and cLf was mostly crystalized into yeast evacuated cells but some protein could still be seen on the surface of packed cells (or may be coming out of cells).
Overall, cLf was successfully packed into S. cerevisiae evacuated cells delivery system and showed antifungal activity against C. albicans at MIC of 2.6 mg/ml that was comparable to antifungal activity of free amphotericin B at MIC of 5 μg/ml or 10.5 μg/ml of evacuated cells delivery system for amphotericin B.
Conclusion
Evacuated S. cerevisiae cells prepared using an optimized sponge-like protocol is a potential drug delivery system for cLf. This system can be developed into topical formulation to treat C. albicans infection.
Author Statements
Author Contributions
Abd El-Baky N and Amara AA: Conceptualization and Methodology; Rezk NMF conducted light microscopic inspection of cells and electrophoresis; Abd El-Baky N and Amara AA performed data curation and analysis; Abd El-Baky N and Amara AA wrote, proofread, revised, and finalized the manuscript. All authors have read and agreed to the published version of the manuscript.
References
- Hu X, Zhang J. Yeast capsules for targeted delivery: the future of nanotherapy? Nanomedicine (Lond). 2017; 12: 955-7.
- Yeh YC, Huang TH, Yang SC, Chen CC, Fang JY. Nano-based drug delivery or targeting to eradicate bacteria for infection mitigation: a review of recent advances. Front Chem. 2020; 8: 286.
- Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021; 20: 101-24.
- Tan Y, Chen L, Li K, Lou B, Liu Y, Liu Z. Yeast as carrier for drug delivery and vaccine construction. J Control Release. 2022; 346: 358-79.
- Salari R, Bazzaz BSF, Rajabi O, Khashyarmanesh Z. New aspects of Saccharomyces cerevisiae as a novel carrier for berberine. Daru. 2013; 21: 73.
- Batah AM, Ahmad TA. The development of ghost vaccines trials. Expert Rev Vaccines. 2020; 19: 549-62.
- Paukner S, Kohl G, Lubitz W. Bacterial ghosts as novel advanced drug delivery systems: antiproliferative activity of loaded doxorubicin in human Caco-2 cells. J Control Release. 2004; 94: 63-74.
- Amara AA, Salem-Bekhit MM, Alanazi FK. Sponge-like: a new protocol for preparing bacterial ghosts. Scientific World Journal. 2013; 2013: 545741.
- Amro AA, Salem-Bekhit MM, Alanazi FK. Plackett-Burman randomization method for bacterial ghosts preparation from E. coli JM109. Saudi Pharm J. 2014; 22: 273-9.
- Alanazi FK, Alsuwyeh AA, Haq N, Salem-Bekhit MM, Al-Dhfyan A, Shakeel F. Vision of bacterial ghosts as drug carriers mandates accepting the effect of cell membrane on drug loading. Drug Dev Ind Pharm. 2020; 46: 1716-25.
- Keshmiri-Neghab H, Goliaei B. Therapeutic potential of gossypol: an overview. Pharm Biol. 2014; 52: 124-8.
- Amara AA. Bacterial and yeast ghosts: E. coli and Saccharomyces cerevisiae preparation as drug delivery model. Int Sci Investig J. 2015; 4: 11-22.
- Yin L, Yuvienco C, Montclare JK. Protein based therapeutic delivery agents: contemporary developments and challenges. Biomaterials. 2017; 134: 91-116.
- Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, et al. Exosomes as drug delivery vehicles for Parkinson’s disease therapy. J Control Release. 2015; 207: 18-30.
- Knudsen KB, Northeved H, Kumar PE, Permin A, Gjetting T, Andresen TL, et al. In vivo toxicity of cationic micelles and liposomes. Nanomedicine. 2015; 11: 467-77.
- Busatto S, Iannotta D, Walker SA, Di Marzio L, Wolfram J. A simple and quick method for loading proteins in extracellular vesicles. Pharmaceuticals (Basel). 2021; 14: 356.
- Kowalczyk P, Kaczynska K, Kleczkowska P, Bukowska-Osko I, Kramkowski K, Sulejczak D. The lactoferrin phenomenon-A Miracle molecule. Molecules. 2022; 27: 2941.
- Fernandes KE, Carter DA. The antifungal activity of lactoferrin and its derived peptides: mechanisms of action and synergy with drugs against fungal pathogens. Front Microbiol. 2017; 8: 2.
- Gruden Š, Poklar Ulrih N. Diverse mechanisms of antimicrobial activities of lactoferrins, lactoferricins, and other lactoferrin-derived peptides. Int J Mol Sci. 2021; 22: 11264.
- Wakabayashi H, Abe S, Okutomi T, Tansho S, Kawase K, Yamaguchi H. Cooperative anti-Candida effects of lactoferrin or its peptides in combination with azole antifungal agents. Microbiol Immunol. 1996; 40: 821-5.
- El-Baky NA. Differential antimicrobial effectiveness of camel lactoferrin-oleic acid and bovine lactoferrin-oleic acid complexes against several pathogens. SOJ Biochem. 2018; 4: 1-9.
- Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika. 1946; 33: 305-25.
- Redwan EM, Tabll A. Camel lactoferrin markedly inhibits hepatitis C virus genotype 4 infection of human peripheral blood leukocytes. J Immunoassay Immunochem. 2007; 28: 267-77.
- Almehdar HA, El-Baky NA, Alhaider AA, Almuhaideb SA, Alhaider AA, Albiheyri RS, et al. Bacteriostatic and bactericidal activities of camel lactoferrins against Salmonella enterica serovar Typhi. Probiotics Antimicrob Proteins. 2020; 12: 18-31.
- Nath L, Laldinchhana CAD, Choudhury AD, Barakoti H, Devi CM. Development and validation of UV-vis spectrophotometric method for estimation of amphotericin B. Res J Pharm Technol. 2020; 13: 55-9.
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976; 72: 248-54.
- Stowe RA, Mayer RP. Efficient screening of process variables. Ind Eng Chem. 1966; 58: 36-40.
- Gutte A. Yeast as carrier for drug delivery system. IJSREM. 2022; 06: 1-7.
- Zhang L, Peng H, Zhang W, Li Y, Liu L, Leng T. Yeast cell wall particle mediated nanotube-RNA delivery system loaded with miR365 antagomir for post-traumatic osteoarthritis theray via oral route. Theranostics. 2020; 10: 8479-93.
- Soto ER, Ostroff GR. Characterization of multilayered nanoparticles encapsulated in yeast cell wall particles for DNA delivery. Bioconjug Chem. 2008; 19: 840-8.
- Garello F, Stefania R, Aime S, Terreno E, Delli Castelli D. Successful entrapping of liposomes in glucan particles: an innovative micron-sized carrier to deliver water-soluble molecules. Mol Pharm. 2014; 11: 3760-5.
- Rotrekl D, Šalamúnová P, Paráková L, Bado O, Saloň I, Štěpánek F, et al. Composites of yeast glucan particles and curcumin lead to improvement of dextran sulfate sodium-induced acute bowel inflammation in rats. Carbohydr Polym. 2021; 252: 117142.
- Hester MM, Lee CK, Abraham A, Khoshkenar P, Ostroff GR, Levitz SM, et al. Protection of mice against experimental cryptococcosis using glucan particle-based vaccines containing novel recombinant antigens. Vaccine. 2020; 38: 620-6.
- Shibasaki S, Aoki W, Nomura T, Miyoshi A, Tafuku S, Sewaki T, et al. An oral vaccine against candidiasis generated by a yeast molecular display system. Pathog Dis. 2013; 69: 262-8.
- Alper RE, Dainko JL, Schlenk F. Properties of yeast cell ghosts obtained by ribonuclease action. J Bacteriol. 1967; 93: 759-65.
- Haidinger W, Mayr UB, Szostak MP, Resch S, Lubitz W. Escherichia coli ghost production by expression of lysis gene E and staphylococcal nuclease. Appl Environ Microbiol. 2003; 69: 6106-13.
- Amara AA. Saccharomyces cerevisiae ghosts using the sponge-like re-reduced protocol. SOJ Biochem. 2015; 2: 4.
- Levin DE. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005; 69: 262-91.
- Cao C, Cao Z, Yu P, Zhao Y. Genome-wide identification for genes involved in sodium dodecyl sulfate toxicity in Saccharomyces cerevisiae. BMC Microbiol. 2020; 20: 34.
- Tukmachev VA, Nedospasova LV, Zaslavskii BIu, Rogozhin SV. Deistvie dodetsilsul’fata natriia na biologicheskie membrany [Effect of sodium dodecyl sulfate on biological membranes]. Biofizika. 1979; 24: 55-60.
- Zhang T, Lei J, Yang H, Xu K, Wang R, Zhang Z. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast. 2011; 28: 795-8.
- Thacker J. Inactivation and mutation of yeast cells by hydrogen peroxide. Mutat Res. 1975; 33: 147-56.
- Picazo C, Molin M. Impact of hydrogen peroxide on protein synthesis in yeast. Antioxidants (Basel). 2021; 10: 952.
- Almehdar HA, Abd El-Baky N, Mattar EH, Albiheyri R, Bamagoos A, Aljaddawi A, et al. Exploring the mechanisms by which camel lactoferrin can kill Salmonella enterica serovar typhimurium and Shigella sonnei. PeerJ. 2023; 11: e14809.