
Research Article
Austin J Biotechnol Bioeng. 2015;2(3): 1044.
Molecular Characterization of Sweet Cherry Genotypes and Rootstocks by using Prunus SSR Sequences
Erogul D* and Cakir B
Department of Horticulture, University of Ege, Turkey
*Corresponding author: Erogul D, Department of Horticulture, University of Ege, Turkey
Received: April 09, 2015; Accepted: May 29, 2015; Published: June 02, 2015
Abstract
Sweet cherry nurseries graft rootstock onto Prunus avium, Prunus cerasus, Prunus mahaleb and hybrids of other Prunus species. In this study, we used eight Simple Sequence Repeat (SSR) primer pairs to describe the genetic variability of the commercial rootstocks CAB6P, MaxMa 60, MaxMa 14, PHL-C, SL 64 and an unknown rootstock widely used in the production of sweet cherries in Turkey and twelve sweet cherry genotypes. These SSR primers are widely used for molecular characterization of Prunus species. Amplification of SSR loci was obtained for all microsatellite primer pairs and the microsatellites generated multiple alleles (42 in total) in the cherry rootstocks. All tested loci were polymorphic, with four to seven (average = 5.3) alleles per locus. The allele size varied from 95 to 206 bp. UCDCH-17 and UCDCH-21 had the lowest number of alleles per locus (four), while PS12A02 had the most (seven). The observed mean heterozygosity value for different loci was 0.79, while the expected heterozygosity was 0.58.
Keywords: Sweet cherry; Microsatellites; Prunus; Rootstock; SSR transferability
Introduction
Cherry (Prunus avium L.) is economically very important temperate fruit in Turkey. Cherries are thought to have originated around the Caspian and Black Seas and currently they are found across mainland Europe and in western Asian areas [1]. Cherry is mainly produced in the Central Anatolia, Marmara region [2] and Aegean region due to the early harvest time and sunshine.
Use of rootstocks is very important in fruit-tree nurseries and the numerous benefits of rootstocks necessitate their use for sweet cherry production. Many different types of rootstocks are used for Prunus species worldwide [3,4] each of which has a specific set of advantages and limitations in adapting to different geographic regions. Rootstocks can influence the performance of sweet cherry varieties and are responsible for water and nutrient uptake, resistance to soilborne pathogens, and tolerance to environmental stressors. Scion– rootstock interactions also affect water relations, plant size, pathogen resistance, environmental stress tolerance, nutrient uptake, plant size, flowering, vigor, survival of fruit buds, fruit set, fruit quality, yield, and leaf gas exchange [5-10].
Over the past decade, intensive evaluation of sweet cherry rootstocks has taken place; sweet cherry rootstocks that can impart a wide range of tree vigor and that are appropriate for different pedo-climatic conditions and cultivation systems are available [11]. Sweet cherries are traditionally cultivated by grafting on generative rootstocks or seedlings of Prunus avium, Prunus mahaleb and sometimes other Prunus species [12,13]. Strong interest in sweet cherry production has led to increased interest in breeding existing rootstocks and in selecting new, more appropriate rootstocks.
Microsatellites are present in many organisms and are found in the genomes of plants and organelles [14,15]. Generally, Simple Sequence Repeats (SSRs) are considered the marker of choice for genetic fingerprinting of fruit trees because of their high levels of polymorphism, high degree of reliability and reproducibility and codominant mode of inheritance [16,17]. These markers have been used widely in plants for cultivar identification [18,19]. Microsatellites are accepted as good genetic markers in sweet cherry [20,21]. Primers that frame SSR markers have been cloned, sequenced and used in peach [18,21-27], cherry [20,21,24,28,29-34], apricot [35-37], almond [38] and other Prunus rootstocks [39-41] for molecular characterization and identification of different varieties. In the present study we verified the high cross-species transferability of four SSR markers obtained from peach, plum and apricot to other Prunus species such as sweet cherry and sweet cherry rootstocks and we discriminated sweet cherry genotypes and sweet cherry rootstocks by using eight SSR markers.
Materials and Methods
Plant materials
Twelve sweet cherry genotypes (Kp1, Kp2, Kp3, Kp4, Kp5, Kp6, Kp7, Kp8, Kp9, Kp10, Kp11, Kp12) and ‘0900 Ziraat’ as a reference which is the most important sweet cherry variety grown in Turkey and six sweet cherry rootstocks (CAB6P, SL 64, MaxMa 14, MaxMa 60, PHL-C and one unknown rootstock) (Table 1) were used for SSR analysis. The commercial rootstocks and ‘0900 Ziraat’ were obtained from a commercial nursery located in Kemalpasa, Izmir (Figure 1). All the Prunus genotypes and unknown rootstock were collected from Kemalpasa, a significant growing area for early sweet cherry production. The unknown rootstock has no compatibility problems and a positive effect on yield. Its relationship to other commercial rootstocks and sweet cherry varieties has also been determined.
Rootstocks
Genetic Origin
CAB 6P
Prunus cerasus L.
SL 64
Prunus mahaleb L.
Maxma 14
Prunus mahaleb L. X Prunus avium L.
Maxma 60
Prunus mahaleb L. x Prunus avium L.
PHL-C
Prunus avium L. x Prunus cerasus L.
Unknown
-
Table 1: Rootstocks and their genetic origin.
Figure 1: Kemalpaşa, Izmir, Turkey.
DNA extraction
Genomic DNA was extracted from young leaf tissue using a DNA easy plant mini kit (Qiagen, Germany) according to the manufacturer’s instructions. And RNase treatment was performed on the eluted DNA samples, and the purity and concentration of the DNA were checked on 1% (w/v) agarose gels and a NanoDrop ND- 1000 spectrophotometer.
SSR marker analysis
Eight previously published primer pairs were used for SSR analysis (Table 2). These primers have been widely used in molecular characterization of Prunus species. Polymerase Chain Reaction (PCR) amplification was performed in a 10-μl reaction volume containing 15 ng of DNA, 5 pmol of each primer, 0.5 mm dNTP, and 0.5 units GoTaq DNA polymerase (Promega, Madison, WI) that included 1.5 mM MgCl2. Forward primers of each primer pair were labeled with WellRED fluorescent dyes D2 (black), D3 (green), and D4 (blue) (Proligo, Paris, France). PCR conditions consisted of an initial cycle of 3 min at 94ºC, followed by 35 cycles of 1 min at 94ºC, 1 min at 55–60ºC, and 2 min at 72ºC, with a final extension at 72ºC for 10 min. PCR products were diluted with sample loading solution in proportion to the quantity of fluorescent dye used in labeling, applied to sequencing reaction according to the manufacturer’s instructions (Genome Lab DNA Standard Kit-400) and followed by electrophoresis with a CEQ 8800XL capillary DNA analysis system (Beckman Coulter, Fullerton, CA). Allele sizes were determined for each SSR locus using Beckman’s CEQ fragment analysis software. The analyses were repeated a minimum of two times to ensure reproducibility of the results.
SSR locus
Sequence (5’-3’)
Species origin
References
CPSCT010
TTG GGT AAA TAC TTT ATC ATT TCC TCC CTG AAT AAG GGT TGT GC
Plum
Mnejja et al. [46]
UCD-CH13
ACC CGC TTA CTC AGC TGA ACT TAG CAC TAA GCC TTT GCT GC
Sweet cherry
Struss et al. [17]
UCD-CH17
TGG ACT TCA CTC ATT TCA GAG A ACT GCA GAG AAT TTC CAC AAC CA
Sweet cherry
Struss et al. [17]
UCD-CH21
TTG TTG ACC ATC GAA TAT GAA G GAA GGT ACA TGG CGT GCC
Sweet cherry
Struss et al. [17]
UCD-CH31
TCC GCT TCT CTG TGA GTG TG CGA TAG TTT CCT TCC CAG ACC
Sweet cherry
Struss et al. [17]
UDAp-401
AAA CCC TAG CCG CCA TAA CT GCT AAA GGC CTT CCG ATA CC
Apricot
Messina et al. [49]
UDP96-005
GTA ACG CTC GCT ACC ACA AA CCT GCA TAT CAC CAC CCA G
Peach
Cipriani et al. [18]
PS12A02
GCCACCAATGGTTCTTCC AGCACCAGATGCACCTGA
Cherry
Downey and Iezzoni [21]
Table 2: Simple sequence repeat primer pairs used in this study to characterize sweet cherry genotypes and rootstocks.
The genetic analysis program ‘IDENTITY’ 1.0 [42] was used in order to calculate the number of alleles, expected and observed heterozygosity (He and Ho, respectively), estimated frequency of null alleles, and the Probability of Genetic Identity per locus (PI) [43]. The results were then converted to a similarity matrix, and a dendrogram was constructed using the Unweighted Pair-Group Method with Arithmetic Mean (UPGMA) [44] with the Numerical Taxonomy and Multiware Analysis System software, version 2.0 [45].
Results and Discussions
SSR analysis
Each of the eight tested primer pairs yielded PCR amplification products in the sweet cherry genotypes and sweet cherry rootstocks and all tested loci were polymorphic (Table 3). All microsatellites generated multiple alleles, with a total of 42 alleles and four to seven alleles per locus (average = 5.3). Allele size range varied from 95 to 206 bp. The lowest number of alleles (four) was found in the microsatellite markers UCDCH-17 and UCDCH-21 and the most (seven) were found in PS12A02 (Table 3). The observed mean Ho value for different loci for all of the rootstocks was 0.79 while the He was 0.58. The Ho was higher than expected for all microsatellites except PS12A02 because of the presence of null alleles, which are alleles that fail to amplify during PCR. The Ho identified in the primer pairs ranged from 0.30 (PS12A02) to 0.95 (UCDCH17 and UDP96005). The He value ranged from 0.53 (PS12A02) to 0.63 (CPSCT10). The marker PS12A02 had the highest information value as reflected in the lowest PI (0.27) value, while the least informative locus was UCDCH17 (PI = 0.52) (Table 3). Null allele frequency was the highest in locus PS12A02 (0.14) and lowest in UCD-CH17 (–0.26). Null alleles are generally referred to as alleles that fail to amplify during the PCR.
Locus
N
Allelic size range (bp)
He
Ho
PI
r
CPSCT010
5
170-184
0.63
0.90
0.35
-0.16
UCD-CH13
5
127-145
0.62
0.85
0.31
-0.14
UCD-CH17
4
180-206
0.55
0.95
0.52
-0.26
UCD-CH21
4
95-119
0.62
0.90
0.36
-0.17
UCD-CH31
6
124-182
0.60
0.60
0.31
0
UDAp-401
6
142-188
0.61
0.90
0.37
-0.18
UDP96-005
5
103-135
0.56
0.95
0.46
-0.24
PS12A02
7
143-185
0.53
0.30
0.27
0.14
Total
42
Average
5.3
0.58
0.79
N: Number of Alleles; Ho: Observed Heterozygosity; He: Expected Heterozygosity; PI: Probability of Identity; r: Null Allele Frequencies.
Table 3: List of genetic parameters obtained with simple sequence repeat used in this study.
Genetic similarity measured within the rootstocks and sweet cherries ranged between 0.25-1.00. The dendrogram generated from the UPGMA cluster analysis based on the Jaccard coefficient of genetic similarity classified all thirteen sweet cherry (twelve sweet cherry genotypes and a reference sweet cherry variety, ‘0900 Ziraat’) and six sweet cherry rootstocks (five commercial sweet cherry rootstocks and one unknown rootstock) into two main groups, which are depicted in Figure 2. The unknown rootstock formed a separate group and displayed a distinct branching pattern (Figure 2).
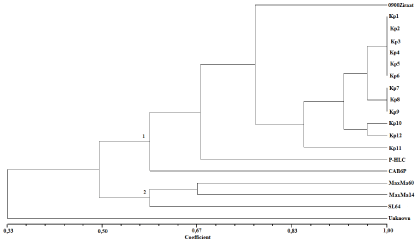
Figure 2: Dendrogram of sweet cherry genotypes and rootstocks based on UPGMA analysis using the genetic similarity matrix generated by the Nei and Li
similarity coefficient after amplification with eight pairs of microsatellite primers.
The first main group included two sweet cherry rootstocks (CAB6P and PHL-C), twelve sweet cherry genotypes and ‘0900 Ziraat’ sweet cherry cultivar. In terms of genetic similarities between the genotypes, the highest genetic similarity (100 %) was observed between the six sweet cherry genotypes (Kp1, Kp2, Kp3, Kp4, Kp5, Kp6) and three sweet cherry genotypes (Kp7, Kp8, Kp9), identified as being synonymous. There was 93.8 % similarity identified between genotypes Kp10 and Kp12. Genotypes of Kp11 shows 81% and 93.8% genetic similarity with Kp genotypes and 68 % similarity with the reference variety (Figure 2) CAB6P (Prunus cerasus L. rootstock clone) and PHL-C (Prunus avium L. × Prunus cerasus L.) rootstocks were separated from others rootstocks and were very closely related to sweet cherry genotypes . Interestingly the rootstock, PHL-C was originated from Prunus avium L., as the cherry genotypes. CAB6P rootstock belongs to the Prunus cerasus rootstock clone and we demonstrated that CAB6P was close to PHL-C rootstock, which is a Prunus avium × Prunus cerasus hybrid clone.
The second main cluster comprises three sweet cherry rootstocks (MaxMa 14, MaxMa 60, SL 64) (Figure 2). Prunus mahaleb L. x Prunus avium L. hybrids, MaxMa 60 and MaxMa 14, grouped in the same cluster. In contrast, SL 64, which is a Prunus mahaleb clone, was closest to the MaxMa clone rootstocks. As shown in Figure 2, surprisingly, SL64 and MaxMa rootstocks were separated from the other rootstocks, PHL-C and CAB6P, originated from Prunus avium x Prunus cerasus and Prunus cerasus clones which were more related to the sweet cherry genotypes (Figure 2).
It was identified that there were 54 alleles with an average of 5.7 alleles per locus in peach and almond by using 27 SSRs including CPSCT10 [46], which was used in the present study. CPSCT10 locus had an average of nine alleles in peach and almond [46], three alleles in wild sweet cherry genotypes [33]; whereas we identified five alleles in sweet cherry genotypes and rootstocks. In the present study, eight loci in sweet cherry genotypes and sweet cherry rootstocks were assayed. The number of alleles per locus ranged from four to seven with an average of 5.3 putative alleles per locus. Previously studies, ten loci assayed in 18 wild sweet cherry genotypes possessed a moderate level of polymorphism, with the number of alleles per locus ranging from three to seven(average 4.6) [33]. Twenty six SSR primers were used in peach and it was detected that there were two to eight (average =4.5) alleles per locus [23]. It was screened that there were 76 sweet cherry genotypes with 34 SSR primer pairs and it was reported that there were 3.7 alleles per locus [29]. Twenty primer pairs were used in order to characterize a wild cherry population and it was found that the number of alleles per locus ranged from four to nine [31]. Thirty three sweet cherry cultivars were used for SSR analysis and it was identified that there were one to six (average = 2.8) alleles per locus [24]. In a survey of 14 sweet cherry cultivars, it was found that there were singlelocus polymorphisms in 19 primer pairs, with two to seven alleles per locus [30].A molecular analysis of 16 wild sweet cherry accessions was done by using ten SSR primers, and it was identified that there were two to six alleles [32]. In another study of sweet cherry, 37 alleles among ten cultivars were obtained by using nine SSR primers [47]. Thirty one sweet cherry cultivars were assessed with 14 SSR primers and it was reported that there were two to eleven alleles (average = 5.3) [48]. In addition, ten SSR primers were used to discriminate Prunus rootstocks and it was demonstrated that the number of alleles ranged from 10 to 20 (average = 13.3) per locus [40].
The UDP96-005 locus developed four alleles in peach [23], five wild sweet cherry genotypes [33] and five in sweet cherry [29,31]. Similarly, the same locus developed five alleles in the present study (Table 3). Fifteen SSR primers were used to characterize sweet cherry cultivars with an average of 3.2 putative alleles per locus and the number of putative alleles ranged from one to five in the tested cherry cultivars; while the number of alleles for UCD-CH13, UCD-CH17, UCD-CH21 and UCD-CH31 loci varied from two to four in sweet cherry [17]. In the present study, we identified four to five alleles per locus. Similarly, the number of alleles for the same locus varied from four to five in wild cherry genotypes [33]. The UDAp-401 locus developed seven alleles in apricot [49], three alleles in wild sweet cherry genotypes [33], while we obtained six alleles sweet cherry genotypes and rootstocks. The PS12A02 locus developed for cherries was the most polymorphic among the eight loci examined in the present study, with the highest effective number of alleles (seven) and the lowest PI value (0.27). The size of the PS12A02 locus varied from 143 to 185 bp. It was also found that PS12A02 was highly informative, with twelve putative alleles (maximum of four per accession) in black sweet cherry and it was determined that this locus was between 150- 178 bp [21]. The same locus was the most polymorphic among the ten loci, and produced the highest effective number of alleles (seven) in wild sweet cherry genotypes [33]. Another study in sweet cherry found 15 alleles per locus for the PS12A02 primer in rootstock [39]; four alleles were found per PS12A02 locus in peaches and five in almond, with a locus size between 175 and 210 bp [38]. Our results indicated that the UCD-CH17 and UCD-CH21 loci were less informative, as they had the lowest allele number (four) and in the case of UCDCH17, a high PI value. The PI value should be >0.05 [50]; all loci examined in this study had PI values >0.05, which indicated that these loci were highly polymorphic for cherry rootstocks. We obtained a similar size range (95-119 bp) for UCD-CH21 with four alleles, while UCD-CH 17 locus was between 180-206 bp. Similarly, [17] it was reported that there were three alleles and a size range of 186-190 bp for UCD-CH 17 and four alleles and a size range of 95-119 bp for UCD-CH21.
Conclusion
In this study, the genetic diversity of twelve sweet cherry genotypes and sweet cherry rootstocks were assessed using eight Prunus SSR primers. Analysis of the genetic structure of the microsatellites supports the effectiveness of microsatellite markers for assessing genetic diversity. These microsatellites, which are widely used for characterization of Prunus species, can be effectively used for sweet cherry rootstocks. Previous studies have shown that the genetic structure in Prunus is well conserved [26,51,52]. Thus, it was determined that SSR markers developed in peaches, sweet cherry and apricot could be utilized in other Prunus species. Various researchers have used SSR markers from different Prunus species to determine genetic characteristics in sweet cherry varieties and rootstocks [21,24,29,31,39,40]. Here, we demonstrated that SSR primers widely used in sweet cherry, peaches, apricot, and plum contained high levels of polymorphisms and are useful for discriminating among sweet cherry rootstocks. In conclusion, our results demonstrated the transferability of SSR markers from cultivated species to wild species in Prunus for the discrimination of genotypes.
References
- 1. Webster AD. The taxonomic classification of sweet and sour cherries and a brief history of their cultivation. Webster AD, Looney NE, editors. In: Cherries: Crop physiology, production and uses, CAB International: Wallingford, Oxon, UK. 1996; 3-24.
- 2. Asma BM. Apricot Growing. Malatya, Turkey. Evin publishers. 2000.
- 3. Rom RC. A new philosophy for peach rootstock development. Fruit Varieties Journal. 1982; 36: 34-37.
- 4. Rom RC. A new generation of peach rootstocks. In: Proceedings of the 43rd National Peach Council Annual Convention of the National Peach Council, Columbia, South Carolina. 1984; 59-68.
- 5. Larsen FE, Higgins SS, RobertFritts JR. Scion/interstock/rootstock effect on sweet cherry yield, tree size and yield efficiency. Scientia Horticulturae. 1987; 33, 237-247.
- 6. Schmitt ER, Duhme F, Schmitt PPS. Water relations in sweet cherries (Prunus avium L.) on sour cherry rootstocks (Prunus cerasus L.) of different compatibility. Scientia Horticulturae. 1989; 39: 189-200.
- 7. Goncalves B, Santos A, Silva AP, Moutinho-Pereira J, Torres-Pereira JMG. Effect of pruning and plant spacing on the growth of cherry rootstocks and their influence on stem water potential of sweet cherry trees. Journal of Horticultural Science and Biotechnology. 2003; 78: 667-672.
- 8. Jiménez S, Garín A, Gogorcena Y, Betrán JA, Moreno MA. Flower and foliar analysis for prognosis of sweet cherry nutrition: influence of different rootstocks. Journal of Plant Nutrition. 2004; 27: 701-712.
- 9. Whiting MD, Lang G, Ophart D. Rootstock and training system affect sweet cherry growth, yield, and fruit quality. HortScience. 2005; 3: 582-586.
- 10. Jiménez S, Pinochet J, Gogorcena Y, Betrán JA, Moreno MA. Influence of different vigour cherry rootstocks on leaves and shoots mineral composition. Scientia Horticulturae. 2007; 112: 73-79.
- 11. Hrotko K. Progress in cherry rootstock research. Acta Horticulturae. 2008; 795: 171-178.
- 12. Schmid PPS, Feucht W. Compability in Prunus avium/Prunus cerasus graftings during the initial phase. III. Isoelectro focusing of proteins, peroxidases and acid phosphatases during union formation. J Hortic Sci. 1985; 60: 311-318.
- 13. Treutter D, Feucht W, Schmid PPS. Polyphenole des Phloems in BeziehungzurInkompatbilita¨ t von Interspezifischen Prunus-Veredlungen (Prunus avium L., Prunus cerasus L.) Flavanone und Flavanole u¨ ber der Veredlungsstelle.Gartenbauwissenschaft. 1986; 51: 77-84.
- 14. Lagercrantz U, Ellegren H, Andersson L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 1993; 21: 1111-1115.
- 15. Wang Z, Weber JL, Zhong G, Tanksley SD. Survey of plant short tandem DNA repeats. Theoretical and Applied Genetics. 1994; 88: 1-6.
- 16. Bouhadida M, Casas AM, Gonzalo MJ, Arus P, Moreno MA´, Gogorcena Y. Molecular characterization and genetic diversity of Prunus rootstocks. Scientia Horticulturae. 2009; 120: 237-245.
- 17. Struss D, Ahmad R, Southwick SM, Boritzki M. Analysis of Sweet Cherry (Prunus avium L.) Cultivars Using SSR and AFLP Markers. Journal of American Society for Horticultural Science. 2003; 128: 904-909.
- 18. Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R. AC/GT and AG/CT Mikrosatellite repeats in peach [Prunus Persica (L) Batsch]: Isolation, characterisation and cross-species amplification in Prunus. Theoretical and Applied Genetics. 1999; 99: 65-72.
- 19. Guilford P, Prakash S, Zhu JM, Rikkerink E. Gardiner S, Bassett H, et al. Microsatellites in MalusX domestica (apple): abundance, polymorphism and cultivar identification. Theoretical and Applied Genetics. 1997; 94: 249-254.
- 20. Cantini C, Iezzoni AF, Lamboy WF, Boritzki M, Struss D. DNA fingerprinting of tetaploid cherry germplasm using SSRs. J Am Soc Hort Sci. 2001; 126: 205-209.
- 21. Downey LD, Iezzoni AF. Polymorphic DNA markers in black cherry are identified using sequences from sweet cherry, peach and sour cherry. J Am Soc Hort Sci. 2000; 125: 76-80.
- 22. Sosinski B, Gannavarapu M, Hager LE, Beck LE, King GJ, Ryder CD, et al. Characterization of microsatellite markers in peach (Prunus persica (L) Basch. Theoretical and Applied Genetics. 2000; 101: 421-428.
- 23. Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT, et al. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome. 2000; 43: 512-520.
- 24. Dirlewanger E, Cosson P, Tavaud M, Aranzana J, Poizat C, Zanetto A, et al. Development of microsatellite markers in peach [ Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry ( Prunus avium L.). Theoretical and Applied Genetics. 2002; 105: 127-138.
- 25. Aranzana MJ, Garcia-Mas J, Carbo J, Arus P. Development and variability analysis of microsatellite markers in peach. Plant Breeding. 2002; 121: 87-92.
- 26. Yamamoto T, Shimada T, Imai T, Yaegaki H, Haji T, Matsuta N, et al. Characterization of morphological traits based on a genetic linkage map in peach. Breeding Science. 2001; 51:271-278.
- 27. Yamamoto T, Mochida K, Imai T, Shi YZ, Ogiware I, Hayashi T. Microsatellite markers in peach [Prunus persica (L.) Batsch] derived from an enriched genomic and cDNA libraries. Molecular Ecology Notes. 2002; 2: 298-301.
- 28. Mohanty A, Martin JP, Aguinanagalde I. Chloroplast DNA study in wild populations and some cultivars of Prunus avium L. Theoretical Applied Genetics. 2001; 103: 112-117.
- 29. Wünsch A, Hormaza JI. Molecular characterisation of sweet cherry (Prunus avium L.) genotypes using peach [Prunus persica (L.) Batsch] SSR sequences. Heredity (Edinb). 2002; 89: 56-63.
- 30. Clarke B, Tobutt KR. Development and characterization of polymorphic microsatellites from Prunus avium ‘Napoleon’. Molecular Ecology Notes. 2003; 3: 578-580.
- 31. Schueler S, Tusch A, Schuster M, Ziegenhagen B. Characterization of microsatellites in wild and sweet cherry (Prunus avium L.)--markers for individual identification and reproductive processes. Genome. 2003; 46: 95-102.
- 32. Vaughan SP, Russel K. Charecterization of novel microsatellites and development of multiplex PCR for large scale population studies in wild cherry, Prunus avium. Molecular Ecology Notes. 2004; 4: 429-431.
- 33. Ercisli S, Agar G, Yildirim N, Duralija B, Vokurka A, Karlidag H. Genetic diversity in wild sweet cherries (Prunus avium) in Turkey revealed by SSR markers. Genet Mol Res. 2011; 10: 1211-1219.
- 34. De Rogatis A, Ferrazzini D, Ducci F, Guerri S, Carnevale S, Elletti P. Genetic variation in Italian wild cherry (Prunus avium L.) as characterized by nSSR markers. Forestry.2013; 86: 391-400.
- 35. Lopes MS, Sefc KM, Laimer M, Machado CA. Identification of microsatellite loci in apricot. Molecular Ecology Notes. 2002; 2: 24-26.
- 36. Hormaza JI. Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theoretical and Applied Genetics. 2002; 104: 321-328.
- 37. Romero C, Pedryc A, Muñoz V, Llácer G, Badenes ML. Genetic diversity of different apricot geographical groups determined by SSR markers. Genome. 2003; 46: 244-252.
- 38. Gomez-Martinez P, Arulsekar S, Potter D, Gradziel TM. An extended interspecific gene pool available to peach and almond breeding as charecterized using Simple Sequence Repeat (SSR) markers. Euphytica. 2003; 131: 313-322.
- 39. Serrano B, Gomez-Aparisi J, Hormaza JI. Molecular fingerprinting of Prunus rootstock using SSRs. J HortiSci Biotechnol. 2002; 77: 368-372.
- 40. Turkoglu Z, Koc A, Ercisli S, Bilgener S, Akbulut M, Yildirim N, et al. Genetic relationships among Prunusrootstocks for sweet cherry (Prunusavium L.) cultivars. Plant Genetic Resources. 2012; 10:101-107.
- 41. Zeinalabedini M, Dezhampour J, Majidian P, Khakzad M, Zanjani BM, Soleimani A, et al. Molecular variability and genetic relationship and structure of Iranian Prunus rootstocks revealed by SSR and AFLP markers.Sci Biotech. 2014; 172: 258-264.
- 42. Wagner HW, Sefc KM. ivCentre for Applied Genetics, Identity 1.0. Vienna: Unersity of Agricultural Science, 1999.
- 43. Paetkau D, Calvert W, Stirling I, Strobeck C. Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol. 1995; 4: 347-354.
- 44. Sneath PH, Sokal RR. Numerical Taxonomy. San Francisco, CA: Freeman, 1973.
- 45. Rohlf FJ. NTYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 1.7. Applied Biostatistics, Setauket, N.Y. 1992.
- 46. Mnejja M, Garcia-Mas J, Howad W, Badenes ML, Arús P. Simple-sequence repeat (SSR) markers of Japanese plum (PrunussalicinaLindl.) are highly polymorphic and transferable to peach and almond. Molecular Ecology Notes. 2004; 4: 163-166.
- 47. AkaKacar Y, Iezzoni AF, Cetiner S. Sweet cherry cultivar identification by using SSR markers. Journal of Biological Sciences. 2005; 5: 616-619.
- 48. Stanys V, Baniulis D, Morkunaite-Haimi S, Siksnianiene JB, Frercks B, Gelvonauskiene D, et al. Characterising the genetic diversity of Lithuanian sweet cherry (Prunus avium L.) cultivars using SSR markers. SciHortic. 2012; 142: 136-142.
- 49. Messina R, Lain O, Marrazzo MT, Cipriani G, Testolin R. New set of microsatellite loci isolated in apricot. Molecular Ecology Notes. 2004; 4: 432-434.
- 50. Sefc KM, Lefort F, Grando MS, Scott KD, Steinkellner H, Thomas MR. Microsatellite markers for grapevine: A state of the art. In: Molecular Biology and Biotechnology of the Grapevine. Kluwer Academic Publishers. 2001; 407-438.
- 51. Joobeur T, Viruel MA, De Vicente MC, Jauregui B, Ballester J, Dettori MT, et al. Construction of a saturated linkage map for Prunus using an almond X peach F2 progeny. Theoretical and Applied Genetics.1998; 97: 1034-1041.
- 52. Wang D, Karle R, Brettin TS, Iezzoni AF. Genetic linkage map in sour cherry using RFLP markers. Theoretical and Applied Genetics. 1998; 97: 1217-1224.