
Special Article- Bioremediation & Biodegradation
Austin J Biotechnol Bioeng. 2015;2(3): 1048.
Development of an Efficient Bioremediation System for Petroleum Hydrocarbon Contaminated Soils Based on Hydrocarbon Degrading Bacteria and Organic Material Control
Dinesh Adhikari, Kiwako S. Araki, Masaki Mukai, Takamitsu Kai, Kenzo Kubota, Taiki Kawagoe, and Motoki Kubo*
Department of Biotechnology, Ritsumeikan University, Japan
*Corresponding author: Motoki Kubo, Department of Biotechnology, Faculty of Life Sciences, Ritsumeikan University, 1-1-1 Nojihigashi, Kusatsu, Shiga 525-8577, Japan
Received: June 25, 2015;Accepted: August 25, 2015; Published: August 27, 2015
Abstract
The efficiency of bioremediation systems can be improved using specific hydrocarbon degrading microorganisms, which necessitates enhancement of number, growth, and activity of these microorganisms. Long-chain cyclic (c)-alkanes are recalcitrant hydrocarbons in soil and water environments. Several long-chain c-alkane degrading bacterial strains have been isolated and characterized, and the numbers of bacteria have also been stimulated, with Rhodococcus erythropolis and Gordonia terrae being most efficient. Degradation efficiency and type of alkane hydroxylase gene were closely related, and bacterial number and hydrocarbon degrading activity were important factors for efficient bioremediation systems. Management of biomass (Total Carbon (TC), Total Nitrogen (TN), and C/N ratio) in the contaminated soil was found to be important to enhancement of the number and activity of Hydrocarbon Degrading Bacteria (HDB). When TC and TN were controlled (TC=20,000 mg/kg, TN=2,000 mg/kg, and C/N ratio=10) using organic materials, the number of HDB was stimulated and maintained for a long time relative to those in soil controlled with inorganic materials, resulting in improved bioremediation efficiency.
Keywords: Long-chain cyclic alkanes; Rhodococcus; Gordonia; Biodegradation; Organic materials
Introduction
Environmental pollution with petroleum hydrocarbons is currently a major global concern that threatens all forms of life in soil, freshwater, groundwater and, marine systems. Physicochemical methods such as incineration, solidification, soil vapor extraction, soil washing, chlorination, and ozonation are used to treat petroleum hydrocarbon contaminated soil [1,2]; however, many of these methods are costly or do not completely remove contaminants [3]. The most common method of hydrocarbon remediation is incineration, which requires a large amount of fossil fuel for burning [4]. However, this process also leads to removal of soil organic matter and microorganisms; accordingly, it requires a long time for the biological activity of soil to return after this type of treatment [5].
Conversely, bioremediation of polluted soils is a promising method for treating a wide range of organic contaminants, including petroleum hydrocarbons. Bioremediation not only effectively removes the soil contaminants, but also regenerates biological activity in the soils. To date, a variety of hydrocarbon-degrading bacteria suitable for bioremediation applications have been isolated, identified, and characterized [6-9].
Enhancement of degradation efficiency is the main challenge to removal of petroleum hydrocarbon from the soil. Activation of indigenous hydrocarbon degrading microorganisms and the use of hydrocarbon degrading microbial strains are the two most common approaches to biodegradation. However, efficient methods for enhancement of biomass and activity of hydrocarbon degrading microorganisms in contaminated soils are needed to improve degradation efficiency.
Here, development of an efficient bioremediation system for petroleum hydrocarbon contaminated soils focusing on characterization of various petroleum hydrocarbon degrading bacteria and control of organic materials in the soil is described.
Bioremediation of Petroleum Hydrocarbon Polluted Soils
Regulations regarding remediation of petroleum hydrocarbon contaminated environments started to be enforced from the early 1980s in several countries, including the United States (1984), the Netherlands (1994), Germany (1998), and Japan (2006) [10]. Several environmentally friendly and cost effective bioremediation systems have been developed for remediation of hydrocarbon polluted soils [3,11,12]. These systems can broadly be grouped into two groups, biostimulation and bioaugmentation.
Biostimulation
The efficiency of bioremediation is affected by the physical, chemical, and microbiological properties of the soil. Since the environmental bacteria in the hydrocarbon contaminated soils are sensitive to damage by the hydrocarbons, the bacterial biomass in the hydrocarbon contaminated soil was less than that in the pristine soil [13]. For effective biostimulation, stimulation and maintaining of indigenous HDB in the contaminated soils are necessary. This is accomplished by generating favorable environmental conditions for degradation of existing hydrocarbons by bacteria via the addition of oxygen, water, and inorganic nutrients [14].
In the presence of large quantities of carbon (e.g., hydrocarbon contamination), other nutrients such as nitrogen (N) and phosphorus (P) tend to be depleted [15]. Accordingly, the addition of nutrients through inorganic and organic materials has shown positive effects on biodegradation of petroleum hydrocarbon in soil [16-21]. It is commonly recommended that the C:N:P ratio of soils be maintained at 100:10:1 by the addition of inorganic materials to enable effective biodegradation of oil contaminated environments [22-25]. However, the effectiveness of biostimulation may depend on several other factors in the soil, including the type of oil component, non-oil organic matter content, and the number and degradation potential of indigenous microorganisms.
Bioaugmentation
Biodegradation is often enhanced by adding previously cultivated specific hydrocarbon degrading microorganisms to the contaminated site. Bioaugmentation is useful in environments with poor reserves of indigenous hydrocarbon degrading microorganisms. Moreover, bioremediation of certain recalcitrant compounds can be made possible through bioaugmentation of efficient hydrocarbon degrading bacteria [26,27]. Several bacterial genera with the ability to degrade petroleum hydrocarbons were isolated and identified from many environments in our previous study, including Gordonia, Acinetobacter, Pseudomonas, and Bacillus [28]. Among these organisms, Rhodococcus and Gordonia are extensively used for bioremediation of petroleum hydrocarbons [10,29,30].
Mechanism for Biodegradation of Petroleum Hydrocarbons
Biodegradation of petroleum hydrocarbon requires specific enzymes and various mechanisms. Several microorganisms can grow by utilizing petroleum hydrocarbon compounds as their sole source of carbon and energy. Following microbial degradation, inert and toxic alkane compounds are converted into less toxic substances that are more easily oxidized by other microorganisms.
The general pathway for biodegradation of hydrocarbons from activation to final metabolism is shown in Figure 1. A variety of chemical and microbial enzymatic reactions must occur for complete degradation of hydrocarbons [31]. Hydrocarbons are insoluble in water; therefore, their degradation requires extracellular saponification by biosurfactants [32-34]. After entering into the microbial cell, several oxidative reactions convert the hydrocarbon via intermediary metabolic pathways such as the tricarboxylic acid cycle [35].
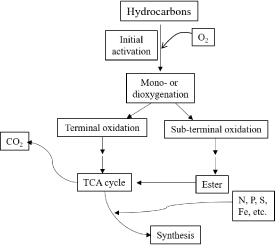
Figure 1: General pathway for hydrocarbon degradation under aerobic
conditions.
The rate of biodegradation depends on the structure of hydrocarbons, with aliphatic hydrocarbons being more easily degraded than aromatic ones (Figure 2). Several enzymes have been found to degrade hydrocarbons. Major microbial enzymes required for initial activation of hydrocarbon degradation under aerobic conditions are listed in Table 1.
Target hydrocarbon
Enzyme for initial activation
References
Short-chain alkanes (C2–C10)
1. Non-heme iron monooxygenase
2. Copper-containing monooxygenase
3. Heme-iron monooxygenase (or cytochrome P450)
[36]
[37]
[38]
Short to medium length chain alkanes (C5–C20)
1. AlkB related alkane hydroxylases
[39]
Long-chain alkanes (>C20)
1. Heme-monooxygenase (P450 type)
2. Non-heme iron monooxygenase (AlkB-related)
3. Flavin-binding monooxygenase (AlmA)
4. Thermophilic flavin-dependent monooxygenase (LadA)
[40]
[41]
[42]
[43,44]
Aromatic hydrocarbons
1. Fe-dioxygenase
2. Fe-monooxygenase
3. Flavin-monooxygenase
[45]
[46]
[47]
Table 1: Microbial enzymes required for initial activation of hydrocarbon degradation under aerobic conditions.
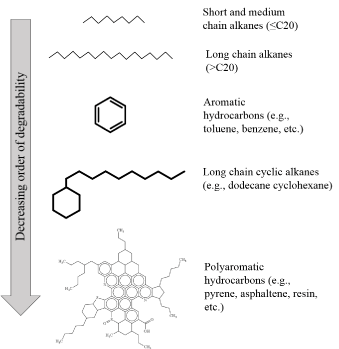
Figure 2: Different types of petroleum hydrocarbons and their relative
degradability.
Isolation and Characterization of Long Chain c-Alkane Degrading Bacteria
Long-chain c-alkanes exist for long periods of time in soil [48]; thus, the isolation and characterization of bacterial strains are important for efficient bioaugmentation of soil contaminated by these compounds.
To enhance the efficiency of bioremediation for long-chain c-alkane hydrocarbon contaminated soils, several bacterial strains were isolated from soils using three types of long chain hydrocarbons (waste car engine oil, base oil, or the c-alkane fraction of the base oil) [28]. More than 400 bacterial strains showed the ability to grow in medium in which the sole carbon source was long chain c-alkanes. Overall, 36 strains showing higher growth ability (OD600≧0.1) was identified based on analysis of 16S rRNA gene sequences. The strains were classified into four groups: actinobacteria (17 strains), gamma-proteobacteria (12 strains), beta-proteobacteria (4 strains), and firmicutes (3 strains). Thirteen actinobacterial strains belonged to the genus Rhodococcus, while the remaining four were Gordonia. Similarly, ten gamma proteobacterial strains belonged to the genus Acinetobacter. The genus Rhodococcus and Gordonia were mainly isolated from medium containing the c-alkane fraction of base oil. A phylogenetic tree of different isolates of the Rhodococcus and Gordonia is presented in Figure 3.
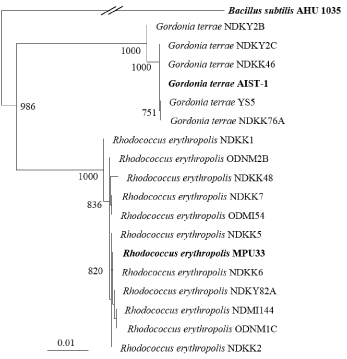
Figure 3: Phylogenetic tree based on alignment of the 16S rRNA gene
sequences (1,460 bp) of long-chain c-alkane degrading genera Rhodococcus
and Gordonia. Bold indicates reference strains.
Degradation of Long Chain c-Alkanes by Rhodococcus and Gordonia
To characterize long-chain c-alkane degradation by Rhodococcus and Gordonia, the relationship between the alkane hydroxylase gene (alkB) sequence and long-chain c-alkane degradation was analyzed (Table 2). R. erythropolis, R. rhodochorus, and R. baikonurensis all contained the alkBR2 type of the alkB gene. Other species of Rhodococcus also carried the alkB gene, but Rhodococcus carrying the alkBR2 type gene showed greater degradation of long-chain c-alkanes than those carrying other types of the alkB gene. Among Gordonia, G. terrae contained the alkBGT type, while all other species had other types of alkB genes. The ability to degrade long-chain c-alkanes also differed among Gordonia based on the alkB gene type, with G. terrae carrying the alkBGT gene showing greater degradation than other types.
Genus
Species
Strain
Alkane hydroxylase gene type
Long-chain c-alkane (DDC) degradation rate*
Rhodococcus
Erythropolis
NDKK6
alkBR2
38.8
Erythropolis
NDKK7
alkBR2
53.5
Erythropolis
ODMI54
alkBR2
68.2
Rhodochorus
NBRC15564
alkBR2
50.6
Baikonurensis
NBRC100611
alkBR2
50.4
Triatomae
NBRC103116
alkBRT
34.0
Maanshanensis
NBRC100610
alkBRM
18.3
Jostii
NBRC16295
alkBRJ
29.8
Koreensis
NBRC100607
alkBRK
6.0
Gordonia
Terrae
NDKY76A
alkBGT
96.5
Terrae
NDKK46
alkBGT
97.8
Terrae
NDKY2B
alkBGT
76.5
Desulfuricans
NBRC100010
alkBGD
84.9
Effuse
NBRC100432
alkBGE
48.2
Araii
NBRC100433
alkBGA
1.9
Otitidis
NBRC100426
alkBGT
0.0
*Degradation rate was analyzed after inoculation of pre-cultured bacterial strains in 100 mL modified SW medium [56] containing 0.1 g DDC followed by incubation at 30°C for 3 days by rotary shaking at 120 rpm.
Table 2: Type of alkB genes in different species of long chain c-alkane degrading Rhodococcus and Gordonia and their rate of Dodecyl Cyclohexane (DDC) degradation.
Previous studies have also shown that Rhodococcus and Gordonia could degrade recalcitrant hydrocarbons such as polyaromatic hydrocarbon compounds [49]. Moreover, these genera could break down even harder to degrade compounds such as phenanthrene, pyrene, and benzo-pyrene [50].
Mechanism of Long Chain c-Alkane Degradation by Rhodoccus and Gordonia
Long-chain hydrocarbons persist in soil for longer than gasoline and light oil [51,52]. Thus, the degradation of long chain c-alkanes by microorganisms is of great importance. Acinetobacter, Rhodococcus, and Gordonia can degrade c-alkanes; however, they utilize different c-alkane degradation pathways [53-55].When dodecylcyclohexane was used as a substrate, Acinetobacter could degrade c-alkanes via co-oxidation of n-alkanes. Conversely, Rhodococcus and Gordonia could degrade c-alkanes without co-oxidation. The putative degradation mechanisms of each genus are shown in Figure 4. The different metabolic pathways of each bacterial genus suggest that a combination of several strains may lead to improved bioremediation of long chain c-alkane contaminated soils.
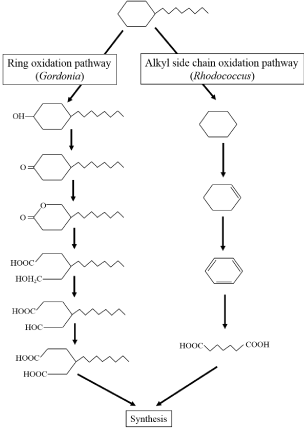
Figure 4: Putative pathways of long-chain c-alkanes degradation by genera
Rhodococcus and Gordonia.
Distribution of Indigenous HDB in the Soil Environment
Activation of indigenous HDB and understanding of their distribution in the soil environment will lead to improve the efficiency of bioremediation. To quantify the indigenous HDB in the soil environment and to investigate the distribution of these bacteria, various soil samples (clay, silt, and sand soils) without hydrocarbon contamination were analyzed. A real-time PCR based method for quantification of hydrocarbon degrading bacteria in the soil has been developed using the alkB gene [56], which encodes a key enzyme involved hydrocarbon degradation [39,41,56]. Since most HDB carry the alkB gene, the distribution of alkB carrying HDB was analyzed to enhance biostimulation.
A total of 23 samples were collected from soils not contaminated with hydrocarbons and analyzed for total bacteria and HDB (Figure 5). HDB in different soils ranged between 3.7×107 and 5.0×108 cells/ g-soil, with an average of 1.3×108 cells/g. The average proportion of HDB was 0.88% of the total bacteria. The presence of hydrocarbondegrading microorganisms in pristine environments has also been reported [57]. Management of the soil environment may enhance the number and activity of these HDB in natural soils, suggesting that enhancing the number of HDB will lead to improved efficiency of bioremediation systems for petroleum hydrocarbon contaminated soils.
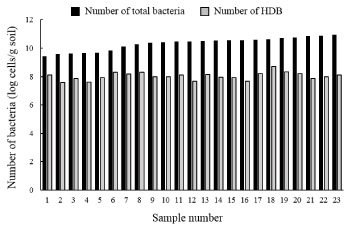
Figure 5: Distribution of total bacteria and HDB in different soil samples
collected from sites not contaminated with petroleum hydrocarbons.
Enhancement of Environmental Bacteria and Material Circulation Activity by Controlling Organic Materials for an Efficient Bioremediation System
Optimization of organic material contents in soil
Enhancements of bacterial number and activity are mandatory for efficient bioremediation of petroleum hydrocarbon contaminated soils. To identify suitable levels of organic materials in soil for enhancing bacterial concentrations and hydrocarbon degrading activity, relationships among the effects of Total Carbon (TC), Total Nitrogen (TN), and C/N ratio on total bacterial biomass and material circulation activity were analyzed in 235 soil samples collected from various fields [58]. More than 75% of the soil samples had C/N ratios of 8 to 25. In soils with this C/N ratio, N circulation gradually increased in accordance with the levels of TC and TN. The number of bacteria was clearly enhanced when TC and TN were present at high levels (Table 3). These results indicate that control of TC, TN, and the C/N ratio in hydrocarbon contaminated soils by organic materials will lead to enhancement of the number and activity of hydrocarbon degrading bacteria.
TC (mg/kg)
TN (mg/kg)
Nitrification activity (point)
Bacterial biomass (× 108 cells/g)
NH4+ oxidation rate (point)
NO2- oxidation rate (point)
Number of samples
≥ 10,000
≥ 1,000
29.3
6.4
35.1
58.8
97
≥ 15,000
≥ 1,500
31.3
6.5
37.9
61.7
61
≥ 20,000
≥ 2,000
36.4
6.7
42.2
68.8
43
≥ 25,000
≥ 2,500
37.7
6.5
41.0
70.0
34
≥ 30,000
≥ 3,000
43.3
7.3
45.8
75.8
27
≥ 40,000
≥ 4,000
47.5
8.2
49.3
85.1
15
≥ 50,000
≥ 5,000
53.9
8.7
56.1
87.9
7
Table 3: Analysis of TC and TN conditions needed for suitable bacterial levels and N circulation activities in soils.
Improvement of bioremediation efficiency by controlling organic materials in soil
Control of organic materials in soil helps to enhance total and hydrocarbon degrading bacterial concentration. Previous studies have shown that improvement of soil biomass via organic manure can enhance the number and activity of indigenous hydrocarbon degrading bacteria [3,11,56]. To enhance the efficiency of bioremediation systems using organic materials, chicken manure was added into the hydrocarbon contaminated soil in a previous study [56]. The concentration of hydrocarbon degrading bacteria was enhanced and maintained for a long time by the addition of organic material (TC: 20,000 mg/kg, TN: 2,000 mg/kg, and C/N ratio 10) (Figure 6). The degradation rate of car engine oil after 28 days was 30.4% when amended with inorganic materials, while it was 47.4% when organic materials were used. Thus, efficiency of petroleum hydrocarbon contaminated soils bioremediation can be enhanced through stimulation of hydrocarbon degrading bacteria using organic material.
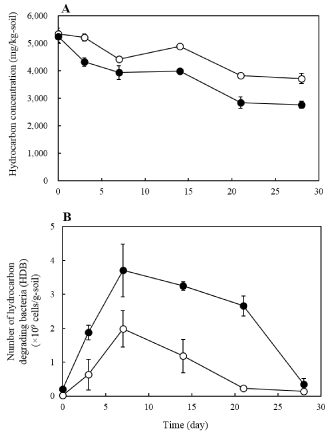
Figure 6: Time course of hydrocarbon concentration (B) and number of HDB
(A) in soils contaminated with petroleum hydrocarbon (car engine oil). Open
circles, biostimulation using inorganic nutrients (SW medium [56]). Filled
circles, biostimulation using organic material (chicken manure). The data are
the average (± standard deviation) of three replications.
Another experiment was conducted to compare the effects of high (TC: 20,000 mg/kg, TN: 1,300 mg/kg, and C/N ratio 15.4) and low (TC: 15,000 mg/kg, TN: 900 mg/kg, and C/N ratio 16.7) levels of TC and TN on bioremediation efficiency of car engine oil contaminated soils. The rate of engine oil degradation was also enhanced when TC and TN were controlled at higher levels (Figure 7). The higher rate of bioremediation of car engine oil corresponded to enhancement and maintenance of the number of R. erythropolis NDKK6 (Figure 8). These findings suggest that proper management of the soil biomass through organic materials is important for enhancing the efficiency of a bioremediation system.
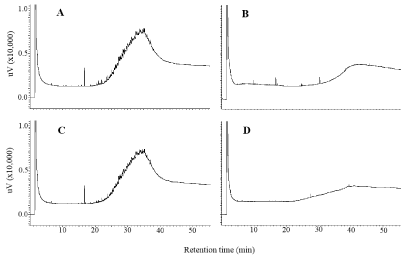
Figure 7: Gas chromatograms of hydrocarbon contents during bioremediation
of soils contaminated with car engine oil. A and B: 0 and 5 weeks after
bioaugmentation of R. erythropolis NDKK6 with high levels of organic matter
(TC: 15,000, TN: 9,300 mg/kg). C and D: 0 and 5 weeks after bioaugmentation
of R. erythropolis NDKK6 with low levels of organic matter (TC: 20,000, TN:
1,300 mg/kg).
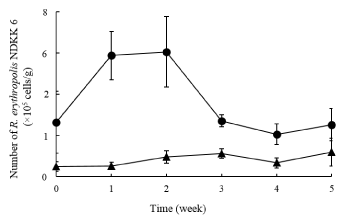
Figure 8: Effects of controlling levels of TC and TN on the concentration of
Rhodococcus erythropolis NDKK6 cells in hydrocarbon contaminated soils
during a bioremediation period of 5 weeks. Filled circles, TC and TN controlled
at high levels (TC: 20,000 mg/kg, TN: 1,300 mg/kg). Filled triangles, TC and
TN controlled at low levels (TC: 15,000 mg/kg, TN: 900 mg/kg). The data are
the averages (± standard deviation) of three replications.
Conclusion
Bioremediation is an emerging approach for treatment of petroleum hydrocarbon contaminated environments. Hydrocarbon degrading bacteria break down and utilize several petroleum hydrocarbons via their specific enzymes. Indigenous HDB exists in a natural soil environment, and enhancement of HDB is very important for an efficient bioremediation system. The use of specific HDB such as Rhodococcus and Gordonia can further enhance bioremediation efficiency. Finally, controlling soil biomass (TC, TN, and C/N ratio) is very important to improving the efficiency of bioremediation systems.
References
- Knight RL, Kadlec RH, Ohlendorf HM. The use of treatment wetlands for petroleum industry effluents. Environ SciTechnol, 1999; 33: 973-980.
- Xu Y, Lu M. Bioremediation of crude oil-contaminated soil: comparison of different biostimulation and bioaugmentation treatments. J Hazard Mater. 2010; 183: 395-401.
- Liu W, Luo Y, Teng Y, Li Z, Ma LQ. Bioremediation of oily sludge-contaminated soil by stimulating indigenous microbes. Environ Geochem Health. 2010; 32: 23-29.
- Matsumiya Y, Kubo M. Bioprocess handbook (in Japanese). NTS Publishing. 2007; 638-652.
- Bárcenas-Moreno G, García-Orenes F, Mataix-Solera J, Mataix-Beneyto J, Bååth E. Soil microbial recolonisation after a fire in a Mediterranean forest. Biol Fertil Soils. 2011; 47: 261-272.
- Pritchard PH, Mueller IJG, Rogers JC, Kremer FV, Glaser JA. Oil spill bioremediation: experiences, lessons and results from the Exxon Valdez oil spill in Alaska. Biodegradation. 1992; 3: 315-335.
- Lal B, Khanna S. Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes odorans. J Appl Bacteriol. 1996; 81: 355-362.
- Vidali M. Bioremediation: an overview. Pure Appl Chem. 2001; 73: 1163-1172.
- Aislabie J, Saul DJ, Foght JM. Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles. 2006; 10: 171-179.
- Hatayama K, Sakihama Y, Matsumiya Y, Kubo M. Construction of efficient bioremediation system for petroleum hydrocarbon-contaminated soils using specific hydrocarbon-degrading bacteria and bacterial biomass monitoring. Soil Contamination Research Trends - New York. Nova Science Publishers, Inc. 2008; 143-160.
- Jørgensen KS, Puustinen J, Suortti AM. Bioremediation of petroleum hydrocarbon-contaminated soil by composting in biopiles. Environ Pollut. 2000; 107: 245-254.
- Hejazi RF, Husain T, Khan FI. Landfarming operation of oily sludge in arid region--human health risk assessment. J Hazard Mater. 2003; 99: 287-302.
- Aoshima H, Kimura A, Shibutani A, Okada C, Matsumiya Y, Kubo M. Evaluation of soil bacterial biomass using environmental DNA extracted by slow-stirring method. Appl Microbiol Biotechnol. 2006; 71: 875-880.
- Couto MN, Monteiro E, Vasconcelos MT. Mesocosm trials of bioremediation of contaminated soil of a petroleum refinery: comparison of natural attenuation, biostimulation and bioaugmentation. Environ Sci Pollut Res Int. 2010; 17: 1339-1346.
- Kvenvolden KA, Cooper CK. Natural seepage of crude oil the marine environment. Geo-Mar Lett. 2003; 23: 140-146.
- Braddock JF, Ruth ML, Catterall PH, Walworth JL, McCarthy KA. Enhancement and inhibition of microbial activity in hydrocarbon-contaminated arctic soils: implications for nutrient-amended bioremediation. Environ Sci Technol.1997; 31: 2078-2084.
- Bundy JG, Paton GI, Campbell CD. Microbial communities in different soil types do not converge after diesel contamination. J Appl Microbiol. 2002; 92: 276-288.
- Delille D, Coulon F, Pelletier E. Effects of temperature warming during a bioremediation study of natural and nutrient-amended hydrocarbon-contaminated sub-Antarctic soils. Cold Regions Sci Technol. 2004; 40: 61-70.
- Margesin R, Labbé D, Schinner F, Greer CW, Whyte LG. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine Alpine soils. Appl Environ Microbiol. 2003; 69: 3085-3092.
- Margesin R, Hämmerle M, Tscherko D. Microbial activity and community composition during bioremediation of diesel-oil-contaminated soil: effects of hydrocarbon concentration, fertilizers, and incubation time. Microb Ecol. 2007; 53: 259-269.
- Xu R, Lau NL, Ng KL, Obbard JP. Application of a slow-release fertilizer for oil bioremediation in beach sediment. J Environ Qual. 2004; 33: 1210-1216.
- Dibble JT, Bartha R. Effect of environmental parameters on the biodegradation of oil sludge. Appl Environ Microbiol. 1979; 37: 729-739.
- USEPA. Land farming, how to evaluate alternative cleanup technologies for underground storage tank sites: a guide for corrective action plan review. Washington DC: United States Environmental Protection Agency. 1994.
- USEPA. Naturally occurring biodegradation as a remedial action option for soil contamination: interim guidance. (Revised) PUBL-SW-515-95. Madison, Wisconsin: Department of Natural Resources and Remedial Response Programme. 1994.
- Leys NM, Bastiaens L, Verstraete W, Springael D. Influence of the carbon/nitrogen/phosphorus ratio on polycyclic aromatic hydrocarbon degradation by Mycobacterium and Sphingomonas in soil. Appl Microbiol Biotechnol. 2005; 66: 726-736.
- Auffret M, Labbé D, Thouand G, Greer CW, Fayolle-Guichard F. Degradation of a mixture of hydrocarbons, gasoline, and diesel oil additives by Rhodococcus aetherivorans and Rhodococcus wratislaviensis. Appl Environ Microbiol. 2009; 75: 7774-7782.
- Fortin NY, Morales M, Nakagawa Y, Focht DD, Deshusses MA. Methyl tert-butyl ether (MTBE) degradation by a microbial consortium. Environ Microbiol. 2001; 3: 407-416.
- Kubota K, Koma D, Matsumiya Y, Chung SY, Kubo M. Phylogenetic analysis of long-chain hydrocarbon-degrading bacteria and evaluation of their hydrocarbon-degradation by the 2, 6-DCPIP assay. Biodegradation. 2008; 19: 749-757.
- Lin TC, Shen FT, Chang JS, Young CC, Arun AB, Lin SY, et al. Hydrocarbon degrading potential of bacteria isolated from oil-contaminated soil. J Taiwan Inst Chem Eng. 2009; 40: 580-582.
- Auffret MD, Yergeau E, Labbé D, Fayolle-Guichard F, Greer CW. Importance of Rhodococcus strains in a bacterial consortium degrading a mixture of hydrocarbons, gasoline, and diesel oil additives revealed by metatranscriptomic analysis. Appl Microbiol Biotechnol. 2015; 99: 2419-2430.
- Sierra-Garcia IN, Oliveira VM. Microbial Hydrocarbon Degradation: Efforts to Understand Biodegradation in Petroleum Reservoirs. Biodegradation Engineer Technol. 2013; 47-72.
- Brusseau ML, Miller RM, Zhang Y, Wang X, Bai GY. Biosurfactant-and cosolvent-enhanced remediation of contaminated media. In: ACS Symposium Series. 1995.
- Bai G, Brusseau ML, Miller RM. Biosurfactant-enhanced removal of residual hydrocarbon from soil. J Contam Hydrol. 1997; 25: 157-170.
- Barkay T, Navon-Venezia S, Ron EZ, Rosenberg E. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Appl Environ Microbiol. 1999; 65: 2697-2702.
- Fritsche W, Hofrichter M. Aerobic degradation by microorganisms. Biotechnology Set, 2nd Edn. 2008; 144-167.
- van Beilen JB, Wubbolts MG, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994; 5: 161-174.
- Hamamura N, Storfa RT, Semprini L, Arp DJ. Diversity in butane monooxygenases among butane-grown bacteria. Appl Environ Microbiol. 1999; 65: 4586-4593.
- Maier T, Förster HH, Asperger O, Hahn U. Molecular characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem Biophys Res Commun. 2001; 286: 652-658.
- vanBeilen JB, Li Z, Duetz WA, Smits TH, Witholt B. Diversity of alkane hydroxylase systems in the environment. Oil Gas Sci Technol. 2003; 58: 427-440.
- vanBeilen JB, Funhoff EG, van Loon A, Just A, Kaysser L, Bouza M, et al. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl Environ Microbiol. 2006; 72: 59-65.
- Nie Y, Liang J, Fang H, Tang YQ, Wu XL. Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl Environ Microbiol. 2011; 77: 7279-7288.
- Maeng JH, Sakai Y, Tani Y, Kato N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J Bacteriol. 1996; 178: 3695-3700.
- Feng L, Wang W, Cheng J, Ren Y, Zhao G, Gao C, et al. Genome and proteome of long-chain alkane degrading Geobacillus thermodenitrificans NG80-2 isolated from a deep-subsurface oil reservoir. Proc Natl Acad Sci U S A. 2007; 104: 5602-5607.
- Li L, Liu X, Yang W, Xu F, Wang W, Feng L, et al. Crystal structure of long-chain alkane monooxygenase (LadA) in complex with coenzyme FMN: unveiling the long-chain alkane hydroxylase. J Mol Biol. 2008; 376: 453-465.
- Larkin MJ, Allen CC, Kulakov LA, Lipscomb DA. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J Bacteriol. 1999; 181: 6200-6204.
- Kim SJ, Kweon O, Jones RC, Edmondson RD, Cerniglia CE. Genomic analysis of polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Biodegradation. 2008; 19: 859-881.
- van Beilen JB, Neuenschwander M, Smits TH, Roth C, Balada SB, Witholt B. Rubredoxins involved in alkane oxidation. J Bacteriol. 2002; 184: 1722-1732.
- Stroud JL, Paton GI, Semple KT. Microbe-aliphatic hydrocarbon interactions in soil: implications for biodegradation and bioremediation. J Appl Microbiol. 2007; 102: 1239-1253.
- Bourguignon N, Isaac P, Alvarez H, Amoroso MJ, Ferrero MA. Enhanced polyaromatic hydrocarbon degradation by adapted cultures of actinomycete strains. J Basic Microbiol. 2014; 54: 1288-1294.
- Pizzul L, delPilar Castillo M, Stenström J. Characterization of selected actinomycetes degrading polyaromatic hydrocarbons in liquid culture and spiked soil. World J Microb Biot. 2006; 22: 745-752.
- Semple KT, Morriss AWJ, Paton GI. Bioavailability of hydrophobic organic contaminants in soils: fundamental concepts and techniques for analysis. Eur J Soil Sci. 2003; 54: 809-818.
- Schmidt MW, Torn MS, Abiven S, Dittmar T, Guggenberger G, Janssens IA, et al. Persistence of soil organic matter as an ecosystem property. Nature. 2011; 478: 49-56.
- Koma D, Sakashita Y, Kubota K, Fujii Y, Hasumi F, Chung SY, et al. Degradation of car engine base oil by Rhodococcus sp. NDKK48 and Gordonia sp. NDKY76A. Biosci Biotechnol Biochem. 2003; 67: 1590-1593.
- Koma D, Sakashita Y, Kubota K, Fujii Y, Hasumi F, Chung SY, et al. Degradation pathways of cyclic alkanes in Rhodococcus sp. NDKK48. Appl Microbiol Biotechnol. 2004; 66: 92-99.
- Koma D, Hasumi F, Chung SY, Kubo M. Biodegradation of n-alkylcyclohexanes by co-oxidation via multiple pathways in Acinetobacter sp. ODDK71. J Biosci Bioeng. 2003; 95: 641-644.
- Fukuhara Y, Horii S, Matsuno T, Matsumiya Y, Mukai M, Kubo M. Distribution of hydrocarbon-degrading bacteria in the soil environment and their contribution to bioremediation. Appl Biochem Biotechnol. 2013; 170: 329-339.
- Greer CW, Whyte LG, Niederberger TD. Microbial communities in hydrocarbon-contaminated temperate, tropical, alpine, and polar soils. In: Handbook of Hydrocarbon and Lipid Microbiology. Springer Berlin Heidelberg. 2010; 2313-2328.
- Adhikari D, Kai T, Mukai M, Araki KS, Kubo M. Proposal for a new soil fertility index (SOFIX) for organic agriculture and construction of a SOFIX database for agricultural fields. Curr TopicsBiotechnol. 2015; 8: 81-91.
Citation: Adhikari D, Araki KS, Mukai M, Kai T, Kubota K, Kawagoe T, et al. Development of an Efficient Bioremediation System for Petroleum Hydrocarbon Contaminated Soils Based on Hydrocarbon Degrading Bacteria and Organic Material Control. Austin J Biotechnol Bioeng. 2015;2(3): 1048. ISSN: 2378-3036