
Review Article
Austin J Biotechnol Bioeng. 2016; 3(1): 1059.
DNA Barcoding: An Effective Technique in Molecular Taxonomy
Purty RS and Chatterjee S*
University School of Biotechnology, Guru Gobind Singh Indraprastha University, India
*Corresponding author: Chatterjee S, University School of Biotechnology, Guru Gobind Singh Indraprastha University, Sector 16C, Dwarka, New Delhi 110078, India
Received: January 26, 2016; Accepted: March 14, 2016; Published: March 16, 2016
Abstract
Global warming is affecting regional climate, ecosystem and diversity array of species by causing physical and biological changes throughout the planet. Therefore, there is a need to develop a technique which can identify organisms and differentiate between very closely related species in order conserve species diversity. Classical taxonomy has accelerated its progress with the adoption of new molecular techniques like DNA barcoding to cope with the huge population of organisms and biodiversity available in this planet. DNA barcoding uses short gene sequences which are well classified portion of the genome. With the advent of high throughput sequencing technology such as Next-Generation Sequencing (NGS) technology the DNA barcoding has become more accurate, fast and reliable in the last decade. The Consortium for the Barcode of Life (CBOL) has given a platform for taxonomists across all the countries to collaborate, identify and preserve the biodiversity across the globe. In this review we summarized the recent advances and developments in the DNA barcoding attempts across animals, plants, bacteria, fungi, viruses and protists. We have also attempted to present the popular tools used in DNA barcoding in a chronological order of their development.
Keywords: DNA barcoding; Bioinformatics tools; Barcoding region; Cytochrome C oxidase (COI); Maturase K (matK); Internal transcribed spacer (ITS)
Introduction
A DNA barcode is one or few relatively short gene sequences present in the genome which is unique enough to identify species. DNA barcoding is a useful tool for taxonomic classification and identification of species by sequencing a very short standardized DNA sequence in a well-defined gene. In this technique, complete information of the species can be obtained from a single specimen irrespective to morphological or life stage characters. It is an effective technique in which extracted DNA from the collected sample is processed following the standard protocol (Figure 1). Identification of the species is carried out by amplifying highly variable region i.e., DNA barcode region of the nuclear, chloroplast or mitochondrial genome using Polymerase Chain Reaction (PCR). Region widely used for DNA barcoding include nuclear DNA (e.g. ITS), chloroplast DNA (e.g. rbcL, trnL-F, matK, psbA, trnH, psbK) and mitochondrial DNA (e.g. COI) (Figure 2). DNA barcodes can be used as a tool for grouping unknown species based on barcode sequence to earlier known species or new species. It can also be used for grouping specimens to known species in those cases where morphologic features are missing or misleading. It can also be used as a supplement to other taxonomic datasets in the process of delimiting species boundaries [1]. The set of DNA barcode markers have been applied to specific taxonomic groups of organisms and are proving invaluable for understanding species boundaries, community ecology, functional trait evolution, trophic interactions and the conservation of biodiversity [2]. The application of NGS technology had expanded the versatility of DNA barcodes across the ‘Tree of Life’, habitats and geographies as new methodologies are explored and developed [3].
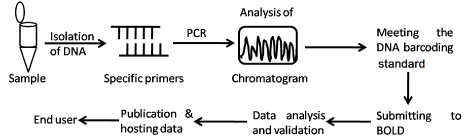
Figure 1: A general DNA barcoding process flow sheet.
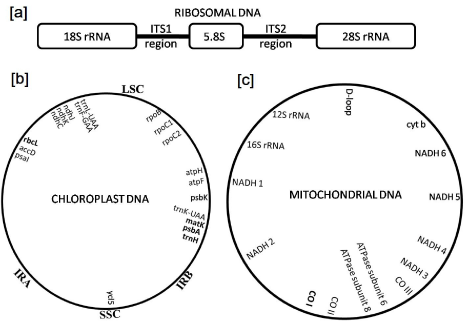
Figure 2: The DNA Barcoding Locus [a] Ribosomal DNA showing ITS region
[b] Chloroplast DNA [c] Mitochondrial DNA.
In order to characterize species, CBOL has selected few genes as ideal for DNA barcoding. Ideally, one gene sequence would be used to identify species in all of the taxa (taxonomic groups) from viruses to plants and animals. However, that ideal gene has not yet been found, so different barcode DNA sequences are used for animals, plants, microbes and viruses. Research using cytochrome c oxidase barcoding techniques on zoological specimens was initiated by Hebert and his group [4]. From 2004, CBOL (currently hosted at https://www. barcodeoflife.org/) started to promote the use of a standardized DNA barcoding approach, consisting of identifying a specimen based on a single universal marker i.e., the DNA barcode sequence. An ideal DNA barcode region or locus should have low intra-specific and high inter-specific divergence (creating a “barcode gap”) and easy to amplify from most or all species in the target group using universal primers. Reference barcodes must be derived from expertly identified vouchers deposited in biological collections with online metadata and validated by available online sequence chromatograms [4].
DNA Barcode of Animals
For animals, the mitochondrial cytochrome C Oxidase I (COI) locus appears to satisfy the desired criteria for most groups [5,6]. This DNA sequence has been found in a mitochondrial gene inherited mainly through the maternal line, which effectively discriminates between most of the animal species (Table 1). Initially, 650 base long segment of the cytochrome C oxidase gene has been elevated to the status of “the barcode of life” for identifying animal species [6]. Later, a 100-base fragment of the original barcode was reported to be effective in identifying archival specimens and potentially useful for all taxa of the eukaryotes [7]. DNA barcoding using COI region is now widely used for molecular evaluation of diversity, as it has good potential for identifying cryptic species and improving our understanding of marine biodiversity [8,9].
Gene/Locus
Organism (s)
Reference (s)
COI
Holothuria edulis
[09]
Leptorhynchoides thecatus
[12]
Pomphorhynchus tereticollis
[13]
Acanthocephalus lucii
[14]
Allolobophora chlorotica
[15]
Polymorphus brevis
[16]
Axiothella constricta, Deosergestes corniculum, Caprella andreae, Microcosmus squamiger, Microcosmus squamiger, Nucula sulcata, Leptoplana tremellaris
[17]
Synecdoche constellate, Bruchomorpha beameri, Cixius nervosus
[18]
Diopatra neapolitana
[19]
Andrena humilis, Andrena fulvida
[20]
Table 1: Gene/locus selected for the study of DNA barcoding in animals.
In animals, mitochondrial DNA occurs as a single double-helical circular molecule containing 13 protein-coding genes, 2 ribosomal genes and several tRNAs (Figure 2c) [10]. Mitochondrial genes are preferred over nuclear genes because mitochondrial genes lack introns, they are generally haploid and exhibit limited recombination [6,11]. Furthermore, each cell has several mitochondria and each mitochondrion contains several such circular DNA molecules and therefore, several complete sets of mitochondrial genes. Thus, when sample tissue is limited, the mitochondrion offers a relatively abundant source of DNA.
DNA Barcode of Plants
DNA barcoding in plants have been a more challenging task than those in animals. Unlike animals, plant mitochondrial genes perform unsatisfactory as a candidate gene for DNA barcoding. The generally low rate of nucleotide substitution in plant mitochondrial genomes precludes the use of COI as a universal plant barcode [21]. However, potential candidates have been reported in the chloroplast genome. The most satisfactory results have come from the gene maturase K (matK) and matK in association with other genes (Table 2). This has been used to resolve the flora of biodiversity hot spots [22]. The matK barcode has been claimed to have discriminated 90 percent of plant species [23].
Gene/Locus
Organism (s)
Barcodes used in combination
Reference (s)
matK
Rhubarb
[29]
Puerariacandollei, Butea superb, Mucunacollettii
[30]
Galpemia spp.
rbcL
[31]
Dendrobium spp.
rbcL
[32]
Angelica spp.
rbcL, ITS ,psbA-trnH
[33]
Rhododendron spp.
rbcL, ITS , psbA-trnH
[34]
753 genera
rbcL, ITS , psbA-trnH
[35]
Lonerica spp.
rbcL, ITS, psbA-trnH, trnL-F
[36]
Solanum spp. & adulterants
rbcL, ITS, psbA-trnH, trnL-F
[37]
Ginseng genus
rbcL, ITS, psbA-trnH, trnL-F, rpoB, rpoC1
[38]
Astragalus spp. & adulterants
rbcL, ITS
[39]
Various medicinal roots
ITS, psbA-trnH, rpoC1
[40]
rbcL
Scutellaria spp. Astragalus spp. and adulterants
matK, psbA-trnH
[41]
Lamiaceae
matK, psbA-trnH
[42]
Various medicinal plants
matK, psbA-trnH
[43]
Sabia spp.
[44]
Pteridophytes
[45]
psbA-trnH
Paris spp. and adulterants
[46]
Senna spp.
[47]
Smilax spp.
[48]
Phyllanthus spp.
[49]
Cistance spp.
[50]
Vitex spp.
matK
[51]
Sideritis spp.
matK
[52]
ITS
Various medicinal plants
[53]
Various medicinal plants
[54]
Boerhavia spp. Astragalus spp. and adulterants
[55]
Sedum spp. Astragalus spp. and adulterants
[56]
Rubus spp.
[57]
Hypericum spp.
[58]
Ochradenus spp.
[59]
Rehmannia spp.
[60]
Medicinal vines (22 genera)
[61]
Dipsacus spp.
[62]
Dendrobium spp.
[25]
Dendrobium spp.
[63]
Paris spp.
[64]
Citrus spp.
[65]
Ruta spp.
[66]
Astragalus spp.
[67]
Meconopsis spp.
[68]
Orobanche spp. and adulterants
[69]
Taraxacumand adulterants
[70]
Table 2: Gene/locus selected for the study of DNA barcoding in plants.
MatK is nested in the group II intron of the chloroplast gene for transfer RNA lysine (trnK), and includes a domain for reverse transcriptase [22]. Group II intron is a class of intron found in rRNA, tRNA and mRNA of organelles in fungi, plants, protists and some mRNA in bacteria. Group II introns are self-splicing in vitro but employ maturase proteins in vivo [24]. Multi-locus markers such as ITS along with matK, rbcL, trnH, etc. have been assumed to be more successful in species identification (Table 2). However, studies to date demonstrated that these are also inadequate for universal plant identification [25,26]. Despite significant recent effort, the development of single-locus barcodes has stalled, placing plant DNA barcoding at a crossroads. Fortunately, developments in DNA sequencing allowing cost-efficient plastid sequencing are driving plant identification into a post-barcode era [27].
The use of two or more chloroplast barcodes has been advocated for the best discrimination in estimating biodiversity, and impressive progress has been made in using chloroplast DNA barcodes for identifying plant species [28].
DNA Barcode of Bacteria
Bacterial 16S rRNA gene is described as an important marker for soil and marine ammonia oxidizing bacteria [71], mountain lakes [72], rice paddy soil microcosms [73] and human clinical samples [74]. So, it is apparent that 16S rRNA gene is highly conserved for each and every species of bacteria and it can be used as a marker for DNA barcode for different species [75]. A microbial diversity picture in this planet is still not clear as many microbes are difficult to culture. It is very crucial to understand microbe association with different environments and their function in those environments. Gene sequences of 16S rRNA from different samples especially environmental samples have reformed our understanding of microbial diversity and it helps us in cataloguing the vast diversity of microorganisms on earth [76]. COI gene was also used to develop the DNA barcode for 22 species pathogen [77]. Smith and his group developed the DNA barcode for Wolbachia, a common endosymbiotic bacterium, using COI gene, and this gene is one of the five multi-locus sequence typing genes which was applied for categorizing Wolbachia [78]. They have found very few overlap with the eukaryotic DNA barcode area. This study corroborates that the COI gene can be a DNA barcode marker for bacteria. Chaperonin-60 (cpn60), also known as GroEL and Hsp60, is a molecular chaperone conserved in bacteria. Conversely, for evaluating the barcoding targets for Archaea including 16S rRNA, type II chaperonin (ortholog of cpn60) was found to be another option [79]. Links and his group suggested that cpn60 can be a common target for bacteria barcode [80]. Some other genes are used for bacterial identification such as rpoB gene which may be used as a barcode marker gene for bacteria [81]. So, we can infer that 16S rRNA, COI gene, and cpn60 can normally be used as markers for developing DNA barcode (Table 3).
Gene/Locus
Organism (s)
Reference (s)
COI
Wolbachia
[78]
rpoB
Streptococcus sp.
[81]
16S
Rickettsia sp., Ehrlichia sp.
[82]
cpn60
Lactobacillus johnsonii, Streptococcus sp.
[80]
tuf
Candidatus phytoplasma sp.
[83]
RIF
Xanthomonas
[84]
gnd
Buchnera sp.
[85]
Table 3: Gene/locus selected for the study of DNA barcoding in bacteria.
DNA Barcode of Fungi
A region of the mitochondrial gene encoding the cytochrome C Oxidase (COI) is the barcode for animals [4,6] and the default marker adopted by the Consortium for the Barcode of Life for all groups of organisms, including fungi [1]. In Oomycota, part of the kingdom Stramenopila historically studied by mycologists, the de facto barcode Internal Transcribed Spacer (ITS) region is suitable for identification, but the default COI marker is more reliable in a few clades of closely related species [86].
COI performs well in Penicillium and other fungi [87], but in few other groups it may not be equally promising, and cloning may often be required [88]. The degenerate primers for many Ascomycota may not be easy to assess as amplification failures may not reflect priming mismatches [89]. Extreme length variation occurs because of multiple introns [87,90], which are not consistently present in a species. Multiple copies of different lengths and variable sequences occur, with identical sequences sometimes shared by several species [89]. Most interestingly, some fungal clades, such as Neocallimastigomycota may lack mitochondria [91]. Most fungi are microscopic and inconspicuous and many are unculturable. Thus robust, universal primers are required to detect a truly representative profile where COI seems to have many challenges from other candidates like ITS [88]. The large subunit of the nuclear ribosomal RNA (LSU), a favoured phylogenetic marker among many mycologists, had virtually no amplification, sequencing, alignment or editing problems and the barcode gap was superior to the Small Subunit of rRNA (SSU). However, across the fungal kingdom, ITS was generally superior to LSU in species discrimination and had a more clearly defined barcode gap [92]. ITS have been reported to perform as a close second to RNA polymerase II largest subunit (RPB1) as the protein-coding marker (Table 4). However, the much higher PCR amplification success rate for ITS may pose a critical difference in its performance as a barcode [88]. It had been reported that all primer sets have a range of biases therefore an appropriate solution may be to use more than one primer combination [93].
Gene/Locus
Organism (s)
Reference (s)
ITS
Fusarium virguliforme
[94]
Colletotrichum sp., Aureobasidium sp., Pseudocercospora sp.
[95]
Gomphus floccosus
Lactarius sp.
Cantharellus cibarius
Tricholoma viridi-olivaceum
Laccaria vinaceo-avellanea, Ramaria maculatipes
[96]
RPB1 (LSU)
Ramaria rubella,
RPB2 (LSU)
Ramaria stuntzii,
18S (SSU)
Gomphus floccosus
Gautieria otthii
Table 4: Gene/locus selected for the study of DNA barcoding in fungus.
DNA Barcode of Virus
Probably, viruses are the most abundant biological creature on earth. It has been estimated that the total number of virus particles is more than 10 times the total number of cells. The molecular diversity of viruses is complicated, as virus molecular diversity of genome is also complex. The “molecular entity” and virus species still remain as a debate for several scientists. The identification and explanation of molecular entities of virus should be the major objective of DNA barcoding. Very few works have been incited to identify the pathogenically important viruses. Therefore, researchers may aim to develop the barcode for the detection or identification of the virus. Recently, few works have been found in this direction [97]. Wei and his group described the k-mer-based barcode image to identify significant pathogenic Human Entero-Viruses (HEVs) [98]. In this process, the condition of 1<k<7 for a fixed k and a genome barcode was described in terms of the k-mer frequency distribution across the whole genome for all combinations of k-mers. Bluetongue Virus (BTV) is an animal virus which affects the different mammals such as cattle, buffalo, sheep, deer, goats, etc [99]. For the detection of BTV, ultrasensitive technique Bio-Barcode Amplification Assay (BCA) method was developed. This method was used for the specific detection of the outer-core protein VP7 of BTV. Though they have produced protein bases bio-barcode, however, signal DNA annealed to DNA strands bound with the gold nanoparticles which were released by heating and characterized by PCR and real-time fluorescence PCR [100]. To detect Avian Influenza Virus (AIV), a fluorescent DNA barcode-based immunoassay was developed based on the application of sandwich immunoassay and fluorophore-tagged oligo-nucleotides as representative barcodes [101]. To understand the viral biodiversity and development of DNA barcode, no marker DNA or RNA has been developed for viruses. However, it is the time to understand the viral biodiversity with DNA barcoding.
DNA Barcode of Protists
Protists are a diverse and loose grouping of disparate eukaryotic microorganisms. They are unicellular, or they are multi-cellular without specialized tissues with relatively simple organization. This simple cellular organization distinguishes the protists from other eukaryotes. They diverged after Archaea and Bacteria evolved but before plants, animals, or fungi appeared on Earth. The Protist Working Group (ProWG), initiated by the Consortium for the Barcode of Life (CBOL) has assessed the efforts to identify the barcode regions across all protist lineages and now introduced a twostep barcoding approach to assess protistan biodiversity. Various protistan DNA barcodes have been proposed (Table 5). The D1–D2 and/or D2–D3 regions at the 5′ end of 28S rDNA have been positively tested in ciliates [102], haptophytes [103], and acantharians [104] and are also promising for diatoms [105]. Zimmermann and his group have also shown that V4 sub-region on the 18S rRNA gene may serve as a potential candidate for barcoding diatoms [106]. Ribosomal Internal Transcribed Spacers (ITS1 and/or ITS2 rDNA), which are the main fungal barcodes, are also commonly utilized in oomycetes [86], chlorarachniophytes [107] and green algae [108]. They have also been suggested for dinoflagellates [109] and diatoms [110] with some reserve [111]. The mitochondrial gene coding for cytochrome C Oxidase (COI), which has been proposed as the universal barcode for animals, also allows morpho-species identification in red [112] and brown [113] algae, dinoflagellates [114], some raphid diatoms [115], Euglyphida [116], lobose naked [117] and shelled [118] amoebae, coccolithophoridhaptophytes [119] and some ciliates [120]. Other group-specific barcodes include the large subunit of the ribulose-1,5-biphosphate carboxylase–oxygenase gene (rbcL) and the chloroplastic 23S rRNA gene for photosynthetic protists [105,121] and spliced leader RNA genes for trypanosomatids [122]. Clearly, the choice of group-specific barcodes is often a question of tradition or ease of use. The studies systematically comparing the resolution power of different protistan DNA barcodes are rare [123].
Gene/Locus
Organism (s)
Reference (s)
ITS
Bigelowiella natans,
Chlorarachnion reptans,
Gymnochlora stellata ,
Lotharella amoeboformis,
Lotharella globosa,
Lotharella oceanic,
Lotharella vacuolata,
Norrisiella sphaerica,
Partenskyella glossopodia,
[107]
COI
Colpoda sp.,
Blepharisma sp.,
Spirostomum ambiguum,
Spirostomum teres,
Stentor coeruleus,
Drepanomonas revolute,
[124]
rbcL
Chaetoceros decipiens sp. ,
Chaetoceros diademasp,
Thalassiosira anguste-lineata,
Thalassiosira pacifica,
[125]
Pseudo-nitzschia delicatissima,
Odontella aurita,
Fragilaria sp.,
[121]
18S
Tabularia fasciculata
[126]
28S
Thalassiosirales,
Chaetocerotales
[125]
Table 5: Gene/locus selected for the study of DNA barcoding in protists.
Tools to Identify DNA Barcode
Innovative computational approaches for DNA barcoding have been developed in the past based on Compensatory Base Changes (CBCs), Operational Taxonomic Units (OTUs), DNA metabarcoding, locus specific tools [127,128], tool for representing barcode symbology, techniques of neural networks, machine learning, data mining [129], composition vector, etc (Table 6). The two new computational methods used in DNA barcoding have been proposed by using barcode sequences of bacteria, archaea, animals, fungi and land plants [130].
Tools
Launch Year
Method
Available at
TaxI
2005
Distance based
axel.meyer@uni-konstanz.de
CBCAnalyzer
2005
Phylogenies based on CBC
https://cbcanalyzer.bioapps.biozentrum.uni-wuerzburg.de/cgi-bin/index.php
4SALE
2006
RNA alignment and editing
https://4sale.bioapps.biozentrum.uni-wuerzburg.de/
CodonCode Aligner
2007
Codon based
https://www.codoncode.com/index.htm
BPSI
2008
Back Propagation (BP) neural networks
zhangab2008@yahoo.com.cn
SAP
2008
Bayesian phylogenetics
https://ib.berkeley.edu/labs/slatkin/munch/StatisticalAssignmentPackage.html
CAOS
2008
Character Based
https://sarkarlab.mbl.edu/CAOS
TaxonGap
2008
Operational Taxonomic Unit (OTU) based
https://www.kermit.ugent.be/software.php?navigatieId=37&categorieId=17
BioBarcode
2009
Sequence based
https://www.asianbarcode.org
BLOG
2009
Data mining approach
https://dmb.iasi.cnr.it/blog-downloads.php
B
2010
Sequence quality and contig overlap
https://www.nybg.org/files/scientists/dlittle/B.html
OFBG
2010
Spp. Discrimination using oligonucleotide frequencies
https://www.nbri.res.in/ofbg.php
OTUbase
2011
Operational Taxonomic Unit (OTU) based
https://www.bioconductor.org/packages/release/bioc/html/OTUbase.html
jMOTU
2011
Multiple Operational Taxonomic Unit (MOTU) based
https://www.jmotu.com-about.com/
Taxonerator
2011
OTU and taxonomy data based
https://www.taxonnerator.com-about.com/
CLOTU
2011
Amplicon and taxa data
https://www.mn.uio.no/ibv/bioportal/
Eco Primers
2011
Barcode markers and primer based
https://www.grenoble.prabi.fr/trac/ecoPrimers
PTIGS-Idit
2011
psbA-trnHIntergenic Spacer (PTIGS) based
https://psba-trnh-plantidit.dnsalias.org
BRONX
2011
Sequence Identification Incorporating Taxonomic Hierarchy
https://www.nybg.org/files/scientists/dlittle/BRONX.html
Spider
2012
Analysis of species identity and evolution
https://spider.r-forge.r-project.org/SpiderWebSite/spider.html
ISHAM
2013
Mycological classification
https://www.isham.org/
LV barcoding
2013
Locality sensitive hashing-based
https://msl.sls.cuhk.edu.hk/vipbarcoding/
Excali BAR
2014
Calculate intra- and interspecific distances
https://datadryad.com/resource/doi:10.5061/dryad.r458n
VIP Barcoding
2014
Vector-based software
https://msl.sls.cuhk.edu.hk/vipbarcoding/
Q-Bank
2015
identification and detection reference database
https://www.q-bank.eu/
obitools package
2015
NGS data based
https://metabarcoding.org/obitools
Table 6: Methods used in various DNA barcoding tools and their web address.
Compensatory Base Changes (CBCs) are mutations of nucleotide at both positions of a paired structural site, while the pairing is still maintained. CBCs reported in rRNA ITS2 have been successfully used for the verification of closely related species. When there is even one CBC at a conserved paired site in the ITS2 secondary structure, they are found to be sexually incompatible [131]. CBCs thus have been successfully used for the discrimination of closely related species. Software based on CBCs is CBC Analyzer [132]. It has been observed that when the number of informative sites are not large, character methods are often less efficient than distance methods [133]. When OTUs with highly unequal evolutionary separation are included in the data set none of the approaches perform well. ‘‘Molecular Operational Taxonomic Units’’ (MOTU) are clusters of sequences derived by grouping the DNA sequences of a conserved gene or gene fragment. The sequence clusters act as representatives of the genomes from which they are derived. A dataset of sequences can be classified into MOTU at a number of different similarity cut-offs, the cut-off value acting as a parameter to the clustering algorithm. Tools like jMOTU and Taxonerator [134], Taxon Gap [135] and CLOTU are based on this concept.
DNA metabarcoding is a process in which environmental sample is used for identifying a number of organisms simultaneously. Environmental sample or eDNA refers to any DNA collected from soil, water or air. It mostly contains degraded DNA and many of the times only short barcodes can be used from it. Therefore, highly conserved and versatile primers are required in metabarcoding studies along with other constraints imposed by this technique. ecoPrimers [136] and OTUbase [137] are tools for DNA metabarcoding studies.
The Back Propagation (BP) neural network approach has been applied for inferring species membership via DNA barcoding. To implement this approach, Zhang and his group have developed a computer program BPSI (BP Species Identification) and demonstrated the power of machine learning in DNA barcoding [138]. This method in combination with bioinformatics especially aimed at identifying species with non-coding barcodes and it is also advantageous compared to their previously proposed BP neural network approach [139]. The method can be applied through the data mining software BLOG (Barcoding with LOGic formulas) [140]. Alignment also becomes the rate limiting step for constructing profile trees for DNA barcoding purposes. Thus unaligned rRNA sequences can be used as barcodes based on the Composition Vector (CV) approach [141] without the need for sequence alignment. The Composition Vector (CV) method is an alignment free approach, especially suitable for non-protein-coding sequences used as barcodes [142]. CVTree is software for constructing phylogeny trees using a CV approach and includes only prokaryotic proteome data.
Limitation of DNA Barcodes
DNA-based species identification depends on distinguishing intra-specific from inter-specific genetic variation. The ranges of these types of variation are unknown and may differ between taxa. It seems difficult to resolve recently diverged species or new species that have arisen through hybridization. There is no universal gene for DNA barcoding, no single gene that is conserved in all domains of life and exhibits enough sequence divergence for species discrimination. The validity of DNA barcoding therefore depends on establishing reference sequences from taxonomically confirmed specimens. This is likely to be a complex process that will involve cooperation among a diverse group of scientists and institutions. Barcode sequences are, in general, short (approx. 500–1000 bp) and this fundamentally limits their utility in resolving deep branches in phylogenies. Some controversy exists over the value of DNA barcoding, largely because of the perception that this new identification method would diminish rather than enhance traditional morphology-based taxonomy, and species determinations based solely on the genetic divergence could result in incorrect species recognition [1]. However, we must keep open the possibility that the barcode sequences per se and their ever increasing taxonomic coverage could become an unprecedented resource for taxonomy, systematic biology and diagnosis, and may be equally useful [143].
Conclusion
As the advantages and limitations of barcoding become apparent, it is clear that taxonomic approaches integrating DNA sequencing, morphology and ecological studies will achieve maximum efficiency at species identification [144]. DNA barcoding may help speed the work of taxonomists and others interested in species identification. The evidence from a number of studies largely confirms the feasibility of such a system [145]. Despite some drawbacks of using DNA barcoding, the reported success of using the barcoding region in distinguishing species from a range of taxa and to reveal cryptic species is remarkable. Efforts should therefore be made to develop nuclear barcodes to complement the barcoding regions that are currently in use.
Acknowledgment
SC and RSP wish to thank Guru Gobind Singh Indraprastha University, New Delhi, India for providing financial and infrastructural support.
References
- Schindel DE, Miller SE. DNA barcoding a useful tool for taxonomists. Nature. 2005; 435: 17.
- Lahaye R, van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA. 2008; 105: 2923-2928.
- Shokralla S, Gibson JF, Nikbakht H, Janzen DH, Hallwachs W, Hajibabaei M. Next-generation DNA barcoding: using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol Ecol Resour. 2014; 14: 892-901.
- Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003; 270: 313-321.
- Das SK, Chatterjee S. Role of DNA barcoding and molecular phylogenetics in biosystematic studies: with special reference to spiders. Proceedings of National Symposium-cum-workshop on Arachnology with reference to Spider: Ecology, Biology & Taxonomy; 2011 October 13- October 15; 27 Solapur, India. Walchand. 2011.
- Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003; 270: S96-99.
- Hajibabaei M, Smith MA, Janzen DH, Rodriguez JJ, Whitfield JB, Hebert PDN. A minimalist barcode can identify a specimen whose DNA is degraded. Mol Ecol Notes. 2006; 6: 959-964.
- Bucklin A, Steinke D, Blanco-Bercial L. DNA barcoding of marine metazoa. Ann Rev Mar Sci. 2011; 3: 471-508.
- Trivedi S, Aloufi AA, Ansari AA, Ghosh SK. Role of DNA barcoding in marine biodiversity assessment and conservation: An update. Saudi J Biol Sci. 2015.
- Alberts B, Johnson A, Lewis J, Raff R, Roberts K, Walter P. The genetic systems of mitochondria and plastids. Molecular Biology of the Cell. 4th Edn. New York: Garland Science. 2002.
- Blaxter ML. The promise of a DNA taxonomy. Philos Trans R Soc Lond B Biol Sci. 2004; 359: 669-679.
- Steinauer ML, Nickol BB, Ortí G. Cryptic speciation and patterns of phenotypic variation of a highly variable acanthocephalan parasite. Mol Ecol. 2007; 16: 4097-4109.
- Perrot-Minnot MJ. Larval morphology, genetic divergence, and contrasting levels of host manipulation between forms of Pomphorhynchus laevis (Acanthocephala). Int J Parasitol. 2004; 34: 45-54.
- Benesh DP, Hasu T, Suomalainen LR, Valtonen ET, Tiirola M. Reliability of mitochondrial DNA in an acanthocephalan: the problem of pseudogenes. Int J Parasitol. 2006; 36: 247-254.
- Dupont L, Lazrek F, Porco D, King RA, Rougerie R, Symondson WOC. New insight into the genetic structure of the Allolobophora chlorotica aggregate in Europe using microsatellite and mitochondrial data. Pedobiologia. 2011; 54: 217-224.
- Alcántar-Escalera FJ, García-Varela M, Vázquez-Domínguez E, Pérez-Ponce de León G. Using DNA barcoding to link cystacanths and adults of the acanthocephalan Polymorphus brevis in central Mexico. Mol Ecol Resour. 2013; 13: 1116-1124.
- Lobo J, Costa PM, Teixeira MA, Ferreira MS, Costa MH, Costa FO. Enhanced primers for amplification of DNA barcodes from a broad range of marine metazoans. BMC Ecol. 2013; 13: 34.
- Gwiazdowski RA, Foottit RG, Maw HE, Hebert PD. The hemiptera (insecta) of Canada: constructing a reference library of DNA barcodes. PLoS One. 2015; 10: e0125635.
- Lobo J, Teixeira MA, Borges LM, Ferreira MS, Hollatz C. Starting a DNA barcode reference library for shallow water polychaetes from the southern European Atlantic coast. Mol Ecol Resour. 2016; 16: 298-313.
- Schmidt S, Schmid-Egger C, Morinière J, Haszprunar G, Hebert PD. DNA barcoding largely supports 250 years of classical taxonomy: identifications for Central European bees (Hymenoptera, Apoideapartim). Mol Ecol Resour. 2015; 15: 985-1000.
- Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One. 2008; 3: e2802.
- Yu J, Xue JH, Zhou SL. New universal matK primers for DNA barcoding angiosperms. J Syst Evol. 2011; 49: 176-181.
- Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011; 6: e19254.
- Kelchner SA. Group II introns as phylogenetic tools: structure, function, and evolutionary constraints. Am J Bot. 2002; 89: 1651-1669.
- Takamiya T, Wongsawad P, Tajima N, Shioda N, Lu JF, Wen CL, et al. Identification of dendrobium species used for herbal medicines based on ribosomal DNA internal transcribed spacer sequence. Biol Pharm Bull. 2011; 34: 779-782.
- Techen N, Parveen I, Pan Z, Khan IA. DNA barcoding of medicinal plant material for identification. Curr Opin Biotechnol. 2014; 25: 103-110.
- Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. Plant DNA barcoding: from gene to genome. Biol Rev Camb Philos Soc. 2015; 90: 157-166.
- Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007; 2: e508.
- Xu G, Wang X, Liu C, Li W, Wei S, Liu Y, et al. Authentication of official Da-huang by sequencing and multiplex allele-specific PCR of a short maturase K gene. Genome. 2013; 56: 109-113.
- Wiriyakarun S, Yodpetch W, Komatsu K, Zhu S, Ruangrungsi N, Sukrong S. Discrimination of the Thai rejuvenating herbs Pueraria candollei (White Kwao Khruea), Butea superba (Red Kwao Khruea), and Mucuna collettii (Black Kwao Khruea) using PCR-RFLP. J Nat Med. 2013; 67: 562-570.
- Sharma A, Folch JL, Cardoso-Taketa A, Lorence A, Villarreal ML. DNA barcoding of the Mexican sedative and anxiolytic plant Galphimia glauca. J Ethnopharmacol. 2012; 144: 371-378.
- Asahina H, Shinozaki J, Masuda K, Morimitsu Y, Satake M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J Nat Med. 2010; 64: 133-138.
- He Y, Hou P, Fan G, Song Z, Arain S, Shu H, et al. Authentication of Angelica anomala Avé-Lall cultivars through DNA barcodes. Mitochondrial DNA. 2012; 23: 100-105.
- Liu Y, Zhang L, Liu Z, Luo K, Chen S, Chen K. Species identification of Rhododendron (Ericaceae) using the chloroplast deoxyribonucleic acid psbA-trnH genetic marker. Pharmacogn Mag. 2012; 8: 29-36.
- Chen S, Yao H, Han J, Liu C, Song J, Shi L, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010; 5: e8613.
- Sun Z, Gao T, Yao H, Shi L, Zhu Y, Chen S. Identification of Lonicera japonica and its related species using the DNA barcoding method. Planta Med. 2011; 77: 301-306.
- Li M, Au KY, Lam H, Cheng L, Jiang RW, But PP, et al. Identification of Baiying (Herba Solani Lyrati) commodity and its toxic substitute Xungufeng (Herba Aristolochiae Mollissimae) using DNA barcoding and chemical profiling techniques. Food Chem. 2012; 135:1653-1658.
- Zuo Y, Chen Z, Kondo K, Funamoto T, Wen J, Zhou S. DNA barcoding of Panax species. Planta Med. 2011; 77: 182-187.
- Guo H, Wang W, Yang N, Guo B, Zhang S, Yang R, et al. DNA barcoding provides distinction between Radix Astragali and its adulterants. Sci China Life Sci. 2010; 53: 992-999.
- Kool A, de Boer HJ, Krüger A, Rydberg A, Abbad A, Björk L, et al. Molecular identification of commercialized medicinal plants in southern Morocco. PLoS One. 2012; 7: e39459.
- Guo X, Wang X, Su W, Zhang G, Zhou R. DNA barcodes for discriminating the medicinal plant Scutellaria baicalensis (Lamiaceae) and its adulterants. Biol Pharm Bull. 2011; 34: 1198-1203.
- Theodoridis S, Stefanaki A, Tezcan M, Aki C, Kokkini S, Vlachonasios KE. DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Ҫeşme-Karaburun Peninsula (Turkey). Mol Ecol Resour. 2012; 12: 620-633.
- Schori M, Showalter AM. DNA barcoding as a means for identifying medicinal plants of Pakistan. Pak J Bot. 2011; 43: 1-4.
- Sui XY, Huang Y, Tan Y, Guo Y, Long CL. Molecular authentication of the ethnomedicinal plant Sabia parviflora and its adulterants by DNA barcoding technique. Planta Med. 2011; 77: 492-496.
- Ma XY, Xie CX, Liu C, Song JY, Yao H, Luo K, et al. Species identification of medicinal pteridophytes by a DNA barcode marker, the chloroplast psbA-trnH intergenic region. Biol Pharm Bull. 2010; 33: 1919-1924.
- Yang Y, Zhai Y, Liu T, Zhang F, Ji Y. Detection of Valeriana jatamansi as an adulterant of medicinal Paris by length variation of chloroplast psbA-trnH region. Planta Med. 2011; 77: 87-91.
- Monkheang P, Sudmoon R, Tanee T, Noikotr K, Bletter N, Chaveerach A. Species diversity, usages, molecular markers and barcode of medicinal Senna species (Fabaceae, Caesalpinioideae) in Thailand. J Med Plant Res. 2011; 5: 6173-6181.
- Kritpetcharat O, Khemtonglang N, Kritpetcharat P, Daduang J, Daduang S, Suwanrungruang K, et al. Using DNA markers and barcoding to solve the common problem of identifying dried medicinal plants with the examples of Smilax and Cissus in Thailand. J Med Plant Res. 2011; 5: 3480-3487.
- Srirama R, Senthilkumar U, Sreejayan N, Ravikanth G, Gurumurthy BR, Shivanna MB, et al. Assessing species admixtures in raw drug trade of Phyllanthus, a hepato-protective plant using molecular tools. J Ethnopharmacol. 2010; 130: 208-215.
- Han JP, Song JY, Liu C, Chen J, Qian J, Zhu YJ, et al. Identification of Cistanche species (Orobanchaceae) based on sequences of the plastid psbA-trnH intergenic region. Yao Xue Xue Bao. 2010; 45: 126-130.
- Phoolcharoen W, Sukrong S. Molecular analysis of Vitex species using candidate DNA barcoding and PCR-RFLP of the matK gene for authentication of Vitex glabrata. Nat Prod Commun. 2013; 8: 125-128.
- Tezcan M, Vlachonasios KE, Aki C. DNA barcoding study on Sideritis trojana Bornm. an endemic medicinal plant of Ida Mountain, Turkey. Fresen Environ Bull. 2010; 19: 1352-1355.
- Gao T, Yao H, Song J, Liu C, Zhu Y, Ma X, et al. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethnopharmacol. 2010; 130: 116-121.
- Mati E, de Boer H. Ethnobotany and trade of medicinal plants in the Qaysari Market, Kurdish Autonomous Region, Iraq. J Ethnopharmacol. 2011; 133: 490-510.
- Selvaraj D, Shanmughanandhan D, Sarma RK, Joseph JC, Srinivasan RV, Ramalingam S. DNA barcode ITS effectively distinguishes the medicinal plant Boerhavia diffusa from its adulterants. Genom Proteom Bioinform. 2012; 10: 364-367.
- Liu MZ, Luo K, Yao H, Liu P. Authentication of Sedum sarmentosum and its adulterants by application of ITS2 sequences. Zhongguo Xiandai Zhongyao. 2011; 13: 29-31.
- Yang JY, Jang SY, Kim HK, Park SJ. Development of a molecular marker to discriminate Korean Rubus species medicinal plants based on the nuclear ribosomal DNA internal transcribed spacer and chloroplast trnL-F intergenic region sequences. J Korean Soc Appl Biol Chem. 2012; 55: 281-289.
- Kazi T, Hussain N, Bremner P, Slater A, Howard C. The application of a DNA-based identification technique to over-the-counter herbal medicines. Fitoterapia. 2013; 87: 27-30.
- Khan S, Al-Qurainy F, Nadeem M, Tarroum M. Development of genetic markers for Ochradenus arabicus (Resedaceae), an endemic medicinal plant of Saudi Arabia. Genet Mol Res. 2012; 11: 1300-1308.
- Kim YS, Ryuk JA, Kom BS. Discrimination of Korean Rehmannia glutinosa from Chinese Rehmannia glutinosa using sequence-characterized amplified region marker. J Korean Soc Appl Biol Chem. 2012; 55: 1-6.
- Liu Z, Zeng X, Yang D, Ren G, Chu G, Yuan Z, et al. Identification of medicinal vines by ITS2 using complementary discrimination methods. J Ethnopharmacol. 2012; 141: 242-249.
- Li D, Wang Z, Liu X, Yuan Y. Identification of the medicinal plant Dipsacus asperoides from three other species in genus Dipsacus (Dipsaceae) by internal transcribed spacer of ribosomal deoxyribonucleic acid (rDNA ITS). J Med Plant Res. 2012; 6: 289-295.
- Chiang CH, Yu TA, Lo SF, Kuo CL, Peng WH, Tsay HS. Molecular authentication of Dendrobium species by multiplex polymerase chain reaction and amplification refractory mutation system analysis. JASHS. 2012; 137: 438-444.
- Liu T, Ji Y. Molecular authentication of the medicinal plant Paris polyphylla Smith var. yunnanensis (Melanthiaceae) and its related species by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). J Med Plant Res. 2012; 6: 1181-1186.
- Wang H, Kim MK, Kim YJ, Lee HN, Jin H, Chen J, et al. Molecular authentication of the Oriental medicines Pericarpium citri reticulatae and Citri unshius pericarpium using SNP markers. Gene. 2012; 494: 92-95.
- Al-Qurainy F, Khan S, Tarroum M, Al-Hemaid FM, Ali MA. Molecular authentication of the medicinal herb Ruta graveolens (Rutaceae) and an adulterant using nuclear and chloroplast DNA markers. Genet Mol Res. 2011; 10: 2806-2816.
- Xiao WL, Motley TJ, Unachukwu UJ, Lau CB, Jiang B, Hong F, et al. Chemical and genetic assessment of variability in commercial Radix Astragali (Astragalus spp.) by ion trap LC-MS and nuclear ribosomal DNA barcoding sequence analyses. J Agric Food Chem. 2011; 59: 1548-1556.
- Li R, Dao Z. Identification of Meconopsis species by a DNA barcode sequence: the nuclear internal transcribed spacer ITS) region of ribosomal deoxyribonucleic acid (DNA). Afr J Biotechnol. 2011; 10: 15805-15807.
- Sun Z, Song J, Yao H, Han J. Molecular identification of Cistanches Herba and its adulterants based on nrITS2 sequence. J Med Plant Res. 2012; 6: 1041-1045.
- Chiang YC, Chang WT, Chen MD, Lai GH, Chen HJ, Chao J, et al. Rapid identification of the medicinal plant Taraxacum formosanum and distinguishing of this plant from its adulterants by ribosomal DNA internal transcribed spacer (ITS) based DNA barcode. Afr J Biotechnol. 2011; 24: 4838-4843.
- Stephen JR, McCaig AE, Smith Z, Prosser JI, Embley TM. Molecular diversity of soil and marine 16S rRNA gene sequences related to beta-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996; 62: 4147-4154.
- Hiorns WD, Methé BA, Nierzwicki-Bauer SA, Zehr JP. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997; 63: 2957-2960.
- Grosskopf R, Janssen PH, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998; 64: 960-969.
- Clarridge JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004; 17: 840-862, table of contents.
- Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007; 45: 2761-2764.
- Lozupone CA, Knight R. Global patterns in bacterial diversity. Proc Natl Acad Sci USA. 2007; 104: 11436-11440.
- Jones YL, Peters SM, Weland C, Ivanova NV, Yancy HF. Potential use of DNA barcodes in regulatory science: identification of the U.S. Food and Drug Administration's "Dirty 22," contributors to the spread of foodborne pathogens. J Food Prot. 2013; 76: 144-149.
- Smith MA, Bertrand C, Crosby K, Eveleigh ES, Fernandez-Triana J, Fisher BL, et al. Wolbachia and DNA barcoding insects: patterns, potential, and problems. PLoS One. 2012; 7: e36514.
- Chaban B, Hill JE. A 'universal' type II chaperonin PCR detection system for the investigation of Archaea in complex microbial communities. ISME J. 2012; 6: 430-439.
- Links MG, Dumonceaux TJ, Hemmingsen SM, Hill JE. The chaperonin-60 universal target is a barcode for bacteria that enables de novo assembly of metagenomic sequence data. PLoS One. 2012; 7: e49755.
- Case RJ, Boucher Y, Dahllöf I, Holmström C, Doolittle WF, Kjelleberg S. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol. 2007; 73: 278-288.
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991; 173: 697-703.
- Makarova O, Contaldo N, Paltrinieri S, Kawube G, Bertaccini A, Nicolaisen M. DNA barcoding for identification of 'Candidatus Phytoplasmas' using a fragment of the elongation factor Tu gene. PLoS One. 2012; 7: e52092.
- Schneider KL, Marrero G, Alvarez AM, Presting GG. Classification of plant associated bacteria using RIF, a computationally derived DNA marker. PLoS One. 2011; 6: e18496.
- Liu L, Huang X, Zhang R, Jiang L, Qiao G. Phylogenetic congruence between Mollitrichosiphum (Aphididae: Greenideinae) and Buchnera indicates insect-bacteria parallel evolution. Syst Entomol. 2013; 38: 81-92.
- Robideau GP, De Cock AW, Coffey MD, Voglmayr H, Brouwer H, Bala K, et al. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resour. 2011; 11: 1002-1011.
- Seifert KA, Samson RA, Dewaard JR, Houbraken J, Lévesque CA, Moncalvo JM, et al. Prospects for fungus identification using COI DNA barcodes, with Penicillium as a test case. Proc Natl Acad Sci USA. 2007; 104: 3901-3906.
- Dentinger BT, Didukh MY, Moncalvo JM. Comparing COI and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina). PLoS One. 2011; 6: e25081.
- Gilmore SR, Gräfenhan T, Louis-Seize G, Seifert KA. Multiple copies of cytochrome oxidase 1 in species of the fungal genus Fusarium. Mol Ecol Resour. 2009; 9: 90-98.
- Vialle A, Feau N, Allaire M, Didukh M, Martin F, Moncalvo JM, et al. Evaluation of mitochondrial genes as DNA barcode for Basidiomycota. Mol Ecol Resour. 2009; 9: 99-113.
- Bullerwell CE, Lang BF. Fungal evolution: the case of the vanishing mitochondrion. Curr Opin Microbiol. 2005; 8: 362-369.
- Seifert KA. Progress towards DNA barcoding of fungi. Mol Ecol Resour. 2009; 9 Suppl s1: 83-89.
- Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol. 2010; 10: 189.
- Gao R, Zhang G. Potential of DNA barcoding for detecting quarantine fungi. Phytopathology. 2013; 103: 1103-1107.
- Abdelfattah A, Li Destri Nicosia MG, Cacciola SO, Droby S, Schena L. Metabarcoding Analysis of Fungal Diversity in the Phyllosphere and Carposphere of Olive (Olea europaea). PLoS One. 2015; 10: e0131069.
- Khaund P, Joshi SR. DNA barcoding of wild edible mushrooms consumed by the ethnic tribes of India. Gene. 2014; 550: 123-130.
- Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nature Comm. 2014; 5: 3075.
- Wei C, Wang G, Chen X, Huang H, Liu B, Xu Y, et al. Identification and typing of human enterovirus: a genomic barcode approach. PLoS One. 2011; 6: e26296.
- Maan S, Maan NS, Nomikou K, Veronesi E, Bachanek-Bankowska K, Belaganahalli MN, et al. Complete genome characterisation of a novel 26th bluetongue virus serotype from Kuwait. PLoS One. 2011; 6: e26147.
- Yin HQ, Jia MX, Shi LJ, Yang S, Zhang LY, Zhang QM, et al. Nanoparticle-based bio-barcode assay for the detection of bluetongue virus. J Virol Methods. 2011; 178: 225-228.
- Cao C, Dhumpa R, Bang DD, Ghavifekr Z, Høgberg J, Wolff A. Detection of avian influenza virus by fluorescent DNA barcode-based immunoassay with sensitivity comparable to PCR. Analyst. 2010; 135: 337-342.
- Gentekaki E, Lynn DH. High-level genetic diversity but no population structure inferred from nuclear and mitochondrial markers of the peritrichous ciliate Carchesium polypinum in the Grand River basin (North America). Appl Environ Microbiol. 2009; 75: 3187-3195.
- Liu H, Probert I, Uitz J, Claustre H, Aris-Brosou S, Frada M, et al. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci USA. 2009; 106: 12803-12808.
- Decelle J, Suzuki N, Mahé F, de Vargas C, Not F. Molecular phylogeny and morphological evolution of the Acantharia (Radiolaria). Protist. 2012; 163: 435-450.
- Hamsher SE, Evans KM, Mann DG, Poulíčková A, Saunders GW. Barcoding diatoms: exploring alternatives to COI-5P. Protist. 2011; 162: 405-422.
- Zimmermann J, Jahn R, Gemeinholzer B. Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene, including new primers and protocols. Org Divers Evol. 2011; 11: 173-192.
- Gile GH, Stern RF, James ER, Keeling PJ. DNA barcoding of Chlorarachniophytes using nucleomorph ITS sequences. J Phycol. 2010; 46: 743-750.
- Coleman AW. The significance of a coincidence between evolutionary landmarks found in mating affinity and a DNA sequence. Protist. 2000; 151: 1-9.
- Litaker RW, Vandersea MW, Kibler SR, Reece KS, Stokes NA, Lutzoni FM, et al. Recognizing Dinoflagellate species using its rDNA sequences. J Phycol. 2007; 43: 344-355.
- Moniz MB, Kaczmarska I. Barcoding of diatoms: nuclear encoded ITS revisited. Protist. 2010; 161: 7-34.
- Mann DG, Sato S, Trobajo R, Vanormelingen P, Souffreau C. DNA barcoding for species identification and discovery in diatoms. Cryptogamie Algologie. 2010; 31: 557-577.
- Saunders GW. Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future applications. Philos Trans R Soc Lond B Biol Sci. 2005; 360: 1879-1888.
- McDevit DC, Saunders GW. On the utility of DNA barcoding for species differentiation among brown macroalgae (Phaeophyceae) including a novel extraction protocol. Phycological Research. 2009; 57: 131-141.
- Stern RF, Horak A, Andrew RL, Coffroth MA, Andersen RA, Küpper FC, et al. Environmental barcoding reveals massive dinoflagellate diversity in marine environments. PLoS One. 2010; 5: e13991.
- Evans KM, Wortley AH, Mann DG. An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist. 2007; 158: 349-364.
- Heger TJ, Pawlowski J, Lara E, Leander BS, Todorov M, Golemansky V, et al. Comparing potential COI and SSU rDNA barcodes for assessing the diversity and phylogenetic relationships of cyphoderiid testate amoebae (Rhizaria: Euglyphida). Protist. 2011; 162: 131-141.
- Nassonova E, Smirnov A, Fahrni J, Pawlowski J. Barcoding amoebae: comparison of SSU, ITS and COI genes as tools for molecular identification of naked lobose amoebae. Protist. 2010; 161: 102-115.
- Kosakyan A, Heger TJ, Leander BS, Todorov M, Mitchell EA, Lara E. COI barcoding of Nebelid testate amoebae (Amoebozoa: Arcellinida): extensive cryptic diversity and redefinition of the Hyalospheniidae Schultze. Protist. 2012; 163: 415-434.
- Hagino K, Bendif ElM, Young JR, Kogame K, Probert I, Takano Y, et al. New evidence for morphological and genetic variation in the cosmopolitan coccolithophore Emiliana huxleyi (Prymnesiophyceae) from the cox1b-ATP4 genes. J Phycol. 2011; 47: 1164-1176.
- Chantangsi C, Lynn DH, Brandl MT, Cole JC, Hetrick N, Ikonomi P. Barcoding ciliates: a comprehensive study of 75 isolates of the genus Tetrahymena. Int J Syst Evol Microbiol. 2007; 57: 2412-2425.
- MacGillivary ML, Kaczmarska I. Survey of the efficacy of a short fragment of the rbcL gene as a supplemental DNA barcode for diatoms. J Eukaryot Microbiol. 2011; 58: 529-536.
- Votýpka J, Maslov DA, Yurchenko V, Jirků M, Kment P, Lun ZR, et al. Probing into the diversity of trypanosomatid flagellates parasitizing insect hosts in South-West China reveals both endemism and global dispersal. Mol Phylogenet Evol. 2010; 54: 243-253.
- Saunders GW, Kucera H. An evaluation of rbcL, tufA, UPA, LSU and ITS as DNA barcode markers for the marine green macroalgae. Cryptogamie, Algologie. 2010; 31: 487-528.
- Strüder-Kypke M, Lynn DL. Comparative analysis of the mitochondrial cytochrome c oxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst Biodiv. 2010; 8: 131-148.
- Hamsher SE, LeGresley MM, Martin JL, Saunders GW. A comparison of morphological and molecular-based surveys to estimate the species richness of Chaetoceros and Thalassiosira (bacillariophyta), in the Bay of Fundy. PLoS One. 2013; 8: e73521.
- Kaczmarska I, Ehrman JM, Moniz MBJ, Davidovich NA. Phenotypic and genetic structure of interbreeding populations of the diatom Tabularia fasciculata (Bacillariophyta). Phycologia. 2009; 48: 391-403.
- Chatterjee S, Das SK. Bioinformatics approach for understanding the molecular mechanisms through motif analysis. Proceedings of National Seminar on Biotechnology for Sustainable Development; HD2, 2012 February 23 - February 24; Kolkata, India: Heritage. 2012.
- Meena SS. Chatterjee S, Aggarwal KK. Structural Modelling of a-subunit of Ring-hydroxylating Dioxygenases (RHDs) from Microbial Sources. Int J Curr Microbiol App Sci. 2015; 4: 740-752.
- Bhargava M, Sharma A. DNA barcoding in plants: evolution and applications of in silico approaches and resources. Mol Phylogenet Evol. 2013; 67: 631-641.
- Tanabe AS, Toju H. Two new computational methods for universal DNA barcoding: a benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS One. 2013; 8: e76910.
- Coleman AW, Vacquier VD. Exploring the phylogenetic utility of ITS sequences for animals: a test case for abalone (Haliotis). J Mol Evol. 2002; 54: 246-257.
- Wolf M, Friedrich J, Dandekar T, Müller T. CBCAnalyzer: inferring phylogenies based on compensatory base changes in RNA secondary structures. In Silico Biol. 2005; 5: 291-294.
- Saitou N, Nei M. The number of nucleotides required to determine the branching order of three species, with special reference to the human-chimpanzee-gorilla divergence. J Mol Evol. 1986; 24: 189-204.
- Jones M, Ghoorah A, Blaxter M. jMOTU and Taxonerator: turning DNA Barcode sequences into annotated operational taxonomic units. PLoS One. 2011; 6: e19259.
- Slabbinck B, Dawyndt P, Martens M, De Vos P, De Baets B. TaxonGap: a visualization tool for intra- and inter-species variation among individual biomarkers. Bioinformatics. 2008; 24: 866-867.
- Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res. 2011; 39: e145.
- Beck D, Settles M, Foster JA. OTUbase: an R infrastructure package for operational taxonomic unit data. Bioinformatics. 2011; 27: 1700-1701.
- Zhang AB, Savolainen P. BPSI2.0: a C/C++ interface program for species identification via DNA barcoding with a BP-neural network by calling the Matlab engine. Mol Ecol Resour. 2009; 9: 104-106.
- Zhang AB, Feng J, Ward RD, Wan P, Gao Q, Wu J, et al. A new method for species identification via protein-coding and non-coding DNA barcodes by combining machine learning with bioinformatic methods. PLoS One. 2012; 7: e30986.
- Bertolazzi P, Felici G, Weitschek E. Learning to classify species with barcodes. BMC Bioinformatics. 2009; 10 Suppl 14: S7.
- Chu KH1, Xu M, Li CP . Rapid DNA barcoding analysis of large datasets using the composition vector method. BMC Bioinformatics. 2009; 10 Suppl 14: S8.
- Qi J, Luo H, Hao B. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004; 32: W45-47.
- Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madrinan S, Petersen G, et al. A proposal for a standardised protocol to barcode all land plants. Taxon. 2007; 56: 295-299.
- Dasmahapatra KK, Mallet J. Taxonomy: DNA barcodes: recent successes and future prospects. Heredity (Edinb). 2006; 97: 254-255.
- Waugh J. DNA barcoding in animal species: progress, potential and pitfalls. Bioessays. 2007; 29: 188-197.