
Research Article
Austin J Biotechnol Bioeng. 2017; 4(1): 1071.
Bioremedial Application of Pseudomonas aeruginosa in Waste Water Treatment
Shah MP*
Industrial Waste Water Research Laboratory, Applied & Environmental Microbiology Lab, Enviro Technology Limited, India
*Corresponding author: Maulin P Shah, Industrial Waste Water Research Laboratory, Applied & Environmental Microbiology Lab, Enviro Technology Limited, Gujarat, India
Received: October 31, 2016; Accepted: March 14, 2017; Published: March 22, 2017
Abstract
Microbial biodegradation of pollutants has intensified in recent years as mankind strives to find sustainable ways to clean up contaminated environments. These biological processes play a major role in the removal of contaminants in the polluted environment. Utilization of catabolic versatility of naturally occurring microorganisms in biodegrading processes is an essential process to degrade or convert such compounds. Recent developments in molecular microbial ecology offer new tools that facilitate molecular analyses of microbial population at contaminated sites. Both conventional and the molecular methods were used in this study to identify the bacteria from different polluted environments. Bacteria were isolated from the dye industry, Common Effluent Treatment Plant & Final Effluent Treatment Plant of Ankleshwar, Gujarat, India. Then, microbial DNA was isolated and amplified with Pseudomonas aeruginosa specific primers. The amplification of 162 bp specific region of catabolic gene of Pseudomonas aeruginosa confirmed the presence of this organism in the contaminated water collected from different climatic regions in Ankleshwar, Gujarat, India.
Keywords: Biodegradation; Polluted water; Pseudomonas aeruginosa
Introduction
One of the major problems that humans are facing is the restoration of the contaminated environment. Textile dyes contribute as the most important environment-polluting agents. Several classes of such contaminants have been synthesized, and still new products are being synthesized now and then. The textile industry is a large water consumer and produces large volumes of contaminated water. One of such examples is the Ankleshwar Industrial Estate, Ankleshwar, and Gujarat, India, which is a seriously industrialized area and produces millions of liters of improperly treated effluents that are released directly without giving proper treatment. Synthetic dyes released into the environment in the form of effluents by textile, leather, food, paper and printing industries cause severe ecological damages. Wastewater resulting from dyeing and finishing processes has an adverse impact in terms of total organic carbon, biological oxygen demand and chemical oxygen demand. Azo dyes are the main constituents of such pollution because of their wide applicability and usages, and therefore, these are present majorly in textile industrial effluents. Moreover their toxicity and resistance to degradation offer great challenge for removal technologies. In many cases the products formed after the degradation of the parent azo dye molecule are more toxic. These products are mainly in aromatic amine form. Azo dyes have been shown to be mutagenic to the human hepatoma cell line where frame shift mutation was observed [1,2]. Induction in the micronuclei formation in human lymphocytes and in HepG2 cells after treatment with azo dye was also observed [3]. The effluent from a dye-processing plant was shown to be responsible for the mutagenic activity detected in a Brazilian river [4]. Methyl red is mutagenic in nature, and most microbial degradation studies reveal the formation of N, N-dimethyl-phenylenediamine, and a toxic and mutagenic aromatic amine [5] that remains untreated in the culture [6]. Acid violet 7 has a significant ability to induce chromosome aberrations, lipid peroxidation and inhibition of acetylcholinesterase. Its toxicity increases extensively after static biodegradation with Pseudomonas putida, due to the corresponding azo reduction metabolites 4'-aminoacetanilide and 5-acetamido-2-amino-1-hydroxy-3, 6- naphthalene disulphonic acid [7]. Therefore azo dyes are the prime attention of the researchers because of their toxicity perspectives. Several physicochemical and microbial methods have been developed for the removal and detoxification of azo dyes. However the developed physicochemical methods present several drawbacks such as high cost, high generation of sludge, high-energy-requiring irradiation methods requires a lot of dissolved O2, etc. [8,9]. Biological methods represent more proper way of textile azo dye removal. Several microorganisms such as algae, yeast, filamentous fungi and bacteria individually or in consortium are shown to degrade the azo dyes in the presence of nutrients [10-15]. However only yeast species Saccharomyces cerevisiae MTCC 463 has been shown to decolorize azo dye in plain distilled water [16]. Rapid industrialization has necessitated the manufacture and use of different chemicals in day to day life [17]. The textile industry is one of them which extensively use synthetic chemicals as dyes. Wastewaters from textile industries pose a threat to the environment, as large amount of chemically different dyes are used. A significant proportion of these dyes enter the environment via wastewater [18]. Approximately 10,000 different dyes and pigments are used industrially and over 0.7 million tons of synthetic dyes are produced annually, worldwide [19]. The aim of this work is to isolate the bacteria from contaminated sites to assess their potential for bioremediation to develop a byproduct useful for bioremediation purposes. Traditional culture dependent methods are insufficient to isolate many microorganisms from the ecological niche [20-22]. Therefore, it is necessary to explore bioremediation agents using modern molecular approach to overcome the limitation factors involved in poor cultivability of microbes (Phoebe et al., 2001). Bioremediation agents using modern molecular approach to overcome the limitation factors involved in poor cultivability of microbes [23].
Materials and Methods
Bacteria and culture
Bacteria were isolated from the different locations of azo dye contaminated water which was collected from the area of industrial zone. The water sample were streaked in nutrient agar medium and incubated at 37oC for 24 hr. Bacterial isolates were selected based on the colour, shape and the morphology of the colonies. To test for the utilization of pollutants by these bacteria, the selected bacterial isolates were grown in enrichment medium containing azo dye (Remazol Black B).
Enrichment
The enrichment culture was prepared by the addition of azo dyes (Remazol Black B) to the nutrient agar broth medium. Selected bacterial isolates were pre cultivated in nutrient broth for overnight. Nutrient agar broth was centrifuged at high speed for 10 min. Pellet was washed in saline water (3%), re-suspended in 4 ml saline and was used as the inoculum at 3% v/v concentration. Nutrient broth medium containing 3% azo dye (Remazol Black B) was used as the sole source of carbon. In addition inoculum (0.3 ml) from each of these selected isolate was added to the nutrient broth and incubated separately on a rotary shaker at 120 rpm for 12 days. The bacterial isolates which grew in the azo dye dilution were selected. The selected strains were then grown in 5 ml of 100% azo dye (Remazol Black B) for 48 hr in the shaker at 120 rpm.
Identification
Microbial properties of the isolates were determined by gram staining and catalase test. Bacteria smear was stained with crystal violet for 2 min followed by gram’s iodine for 1 min and then safranin for 1 min. Catalase activity of the bacterial isolates were tested with 3% H2O2. Identification of isolates present in different sources of wastewater was also performed by DNA analysis.
Identification by DNA based methods
Bacterial cultures isolated from industrial effluent and wastewater collected from contaminated water with dye industry, common effluent treatment plant, final effluent treatment plant from two different climatic zones, especially the wet zone (South GIDC of Ankleshwar) and the intermediate zone (Central GIDC of Ankleshwar) were used for the DNA extraction. For each DNA extraction, two different protocols were used. In the first protocol, DNA was extracted from the bacterial culture obtained from the wastewater contaminated with the dye containing effluent. The second protocol was performed based on the method described for DNA extraction directly from wastewater [24].
Genomic DNA extraction from bacteria
In the first protocol, the lysis step was performed by adding 10 μl of 1% SDS to 800 μl of bacterial suspension and a gentle rotation was done for approximately 5 min. Then the bacterial suspension was kept in a water bath at 65°C for 30 min and left to cool to room temperature. Stirring rod was placed into the lysed bacterial suspension and ice cold 100% ethanol was added slowly down the stirring rod and rod was rotated for 5 min. Rod with bacterial DNA was immersed in 70% ethanol for 2 min. Then DNA was dissolved in 300 μl of TE buffer. Equal volume of chloroform was added to the DNA sample and mixed well. Mixture was centrifuged at high speed for 1 min at room temperature. Aqueous phase was transferred into a fresh tube. This procedure was repeated twice to separate the aqueous phase. DNA was precipitated by addition of 100% ethanol and centrifugation. Ethanol was removed and pellet was air-dried. DNA was dissolved in 20 μl of TE buffer and was stored at -20°C. The second protocol was performed based on the method described for DNA extraction directly from wastewater [24] (see below). For visualizing the DNA extracts, each extract was electrophoresed on 0.8% agarose gel in 1xTBE buffer, stained with ethidium bromide and examined Under Ultraviolet (UV) light.
Microbial DNA extraction from wastewater
In the second protocol, wastewater obtained from industrial waste was enriched with glucose (10 g/L) by keeping overnight in the shaker and then DNA was directly extracted by the modified direct extraction method. The wastewater sample was centrifuged to pellet down the cells at high speed for 15 min. This was performed three times to pool the pellet. The pellet was washed twice with wash buffer (50 mMTrisHCl, pH 8.00, 5mM EDTA, pH 8.00) before lysis. The pellet was dissolved in 500 μl lysis buffer (100 mMTrisHCl, pH 8.00, 100 mM EDTA, pH 8.00, 1.5 M NaCl) and centrifuged at high speed for 15 min. Supernatant was separated and 75 μl NaOAC and 500 μl of ice cold isopropanol were added and centrifuged at high speed for 15 min. Pellet was washed with 70% ethanol. Ethanol was removed and pellet was air-dried until ethanol was evaporated. Then the pellet was resuspended in 25 μl of deionized water. Extracted DNA was electrophoresed on 0.8% agarose gel containing ethidium bromide and visualized under UV light.
Test organism
To screen for the presence of the bioremediation agent in contaminated water in Ankleshwar, Gujarat, India specific primers were designed and used to identify the bacteria Pseudomonas aeruginosaas it is a common bacteria having biodegrading ability, present in all types of waste in all part of the world [25].
PCR Amplification
A region 162 bp from the catabolic gene of dimethyl glycine was amplified using the forward primer and F 5’GAACGTGCTGGTCTACGACA3’ and the reverse primerR5’GG GATACATGCTGCGGTAGT3’. Each 20 μl PCR mixture contained 40 ngDNA, 200 μM dNTPs, 0.7 μM each of two opposing primers, 1X PCR buffer, 2.5 mM MgCl2 and 0.8 units of Taq polymerase. The amplification cycle consisted of an initial denaturation step of 5 min at 94°C followed by 30 cycles of 1 min at 94°C, 1 min at 58°C, 1 min at 72°C and the final extension step of 10 min at 72°C was included. Final holding temperature was 4°C. Template DNA was omitted from the reaction mixture for the negative control. Amplified PCR products were electrophoresis on 1% agarose gel containing ethidium bromide and visualized under ultraviolet light.
Results and Discussion
Isolation and characterization of bacteria from industrial effluent
Bacteria isolated from the industrial effluent by enrichment culture with azo dye (Remazo Black B) were screened for the dye degrading ability. The isolates that could grow on liquid medium with higher concentration of dye (100%) were selected as the most efficient isolates for the degradation of azo dye (data not shown). Most of these bacterial isolates screened for this study, were white in color and appeared bluish in color under the direct light and negative for gram staining and positive for the catalase test. Most aerobic organism makes catalase. Based on its morphological and biochemical properties, these isolates were identified as gram negative and aerobic.
They found to be characteristics of Pseudomonas aeruginosa. In order to identify this bioremediation organism in contaminated sites from different climatic zones and from different types of wastewater, the microbial populations in wastewater were screened by DNA based techniques. Therefore, genomic DNA was first isolated and then subjected to PCR amplification using species specific primers. DNA extracted from bacterial culture by the first protocols provided a good yield of DNA which could be used in PCR amplification (Figure 1). Direct lysis for DNA extraction by optimization with glucose proved to be very efficient in providing large amounts of DNA from environmental samples (Figure 2) while DNA extraction without optimization was not efficient. All extraction products obtained by different protocols could be amplified with the primers specific for Pseudomonas aeruginosa. Satisfactory amplified products were obtained indicating that the DNA was good in quality in all the DNA preparations and the DNA samples were free of PCR inhibitors.
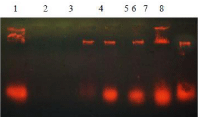
Figure 1: Extraction of Genomic DNA from Bacteria using Protocol 1. Line
1-8: DNA iso1ated from different Bacterial colonies.

Figure 2: Extraction of Microbial DNA from Wastewater. Lane 1: DNA directly
isolated from waste water; Lane 2: DNA isolated from optimized waste water
Samples (Lane 2: Azo dye containing effluent); Lane 3: Common effluent
treatment plant; Lane 4: Final effluent treatment plant; Lane 5: Industrial
waste; Lane 6: Negative control.
PCR Amplification of catabolic gene fragment
PCR amplification was performed to detect the 162 bp region of catabolic gene in Pseudomonas aeruginosa using specific primers. The amplified band was observed between 564 bp and 125 bp region of the lambda ladder, confirming the presence of expected PCR product (Figure 3). The absence of band in negative control explained the reliability of the PCR reaction. However further confirmation is recommended. This could be possible by sequencing the amplified product.
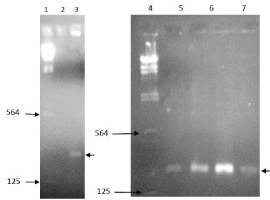
Figure 3: PCR amplification of catabolic gene fragment from environmental
samples Lane 1&4: Lambda DNA ladder; Lane 2: Negative control; Lane 3&7:
Dye containing Effluent; Lane 6: Common effluent treatment plant; Lane 8:
Final effluent treatment plant.
Conclusion
Bacteria isolated from industrial effluent were able to grow and degrade azo dye effectively. These bacterial isolates were identified as gram negative and aerobic. DNA extracted from both bacterial culture and directly from wastewater was good in quality and suitable for PCR amplification. However, DNA extracted directly from wastewater gave high yield of DNA when wastewater was enriched with glucose. The bacterium Pseudomonas aeruginosawas identified from various types of contaminated sites in different parts of the waste water treatment plant of Ankleshwar, Gujarat, India. Further confirmation is required for the detection of this organism. This study should be extended to identify the potential bioremediation organism from other regions of the country.
References
- Ferraz ERA, Grando MD, Oliveira DP. The azo dye Disperse Orange 1 induces DNA damage and cytotoxic effects but does not cause ecotoxic effects in Daphnia similis and Vibrio fischeri. J Hazard Mater. 2011; 192: 628-633.
- Ferraz ERA, Umbuzeiro GAG, Caloto-Oliveira A, Chequer FMD, Zanoni MVB, Dorta DJ, et al. Differential toxicity of Disperse Red 1 and Disperse Red 13 in the Ames test, HepG2 cytotoxicity assay, and Daphnia acute toxicity test. Environ Toxicol. 2011; 26: 489-497.
- Chequer FMD, Angeli F, Ferraz ERA, Tsuboy M, Marcarini J, Mantovani M, et al. The azo dyes Disperse Red 1 and Disperse Orange 1 increase the micronuclei frequencies in human lymphocytes and in HepG2 cells. Mutat Res. 2009; 676: 83-86.
- Umbuzeiro G, Freeman H, Warren S, Oliveira D, Terao Y, Watanabe T, et al. The contribution of azo dyes to the mutagenic activity of the Cristais River. Chemosphere. 2005; 60: 55-64.
- Wong P, Yuen P. Decolourization and biodegradation of N,N'-dimethyl-p-phenylenediamine by Klebsiella pneumoniae RS-13 and Acetobacter liquefaciens S-1. Applied Microbiology. 1998; 5: 79-87.
- Ayed L, Mahdhi A, Cheref A, Bakhrouf A. Decolorization and degradation of azo dye Methyl Red by an isolated Sphingomonas paucimobilis: Biotoxicity and metabolites characterization. Desalination. 2011; 274: 272-277.
- Mansour HB, Ayed-Ajmi Y, Mosrati R, Corroler D, Ghedira K, Barillier D, et al. Acid violet 7 and its biodegradation products induce chromosome aberrations, lipid peroxidation, and cholinesterase inhibition in mouse bone marrow. Environ Sci Pollut Res Int. 2010; 17: 1371-1378.
- Robinson T, McMullan G, Marchant R, Nigam P. Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresource Technology. 2001; 77: 247-255.
- Forgacs E, Cserhati T, Oros G. Removal of synthetic dyes from wastewaters: a review. Environ Int. 2004; 30: 953-971.
- Asgher M, Bhatti HN, Ashraf M, Legge R. Recent developments in biodegradation of industrial pollutants by white rot fungi and their enzyme system. Biodegradation. 2008; 19: 771-783.
- 11El-Sheekh, MM, Gharieb, MM, Abou-El-Souod G. Biodegradation of dyes by some green algae and cyanobacteria. International Biodeterioration and Biodegradation. 2009; 63: 699-704.
- Lin Y, Leu J. Kinetics of reactive azo-dye decolorization by Pseudomonas luteola in a biological activated carbon process. Biochemical Engineering Journal. 2008; 39: 457-467.
- Tan L, Qu Y, Zhou J, Li A, Gou M. Identification and characteristics of a novel salt-tolerant Exiguobacterium sp. for azo dyes decolorization . Appl Biochem Biotechnol. 2009; 159: 728-738.
- Patil PS, Phugare, SS Jadhav, SB, Jadhav JP. Communal action of microbial cultures for Red HE3B degradation. Journal of Hazardous Materials. 2010; 181: 263-270.
- Yu Z, Wen X. Screening and identification of yeasts for decolorizing synthetic dyes in industrial wastewater. International Biodeterioration and Biodegradation. 2005; 56: 109-114.
- Jadhav JP, Parshetti, GK, Kalme SD, Govindwar SP. Decolourization of azo dye methyl red by Saccharomyces cerevisiae MTCC 463. Chemosphere. 2007; 68: 394-400.
- Shah Mp, Patel KA, Nair SS, Darji AM. Optimization of Environmental Parameters on Microbial Degradation of Reactive Black Dye. Journal of Bioremediation & Biodegradation. 2013; 4: 3.
- dye Reactive Red by Bacillus spp ETL. Journal of Bioremediation & Biodegradation. 1982; 4: 3.
- Maulin, P Shah, Kavita A Patel, Sunu S Nair, Darji AM. Microbial degradation of Textile Dye (Remazol Black B) by Bacillus spp ETL-2012. Journal of Bioremediation & Biodegradation. 2013; 4: 2.
- Hugenholtz P, Goebel BM, Pace NR. “Impact of culture independent studies on the emerging phylogenetic view of bacterial diversity” Journal of Bacteriology. 1998; 180: 4765-4774.
- Martin L. DNA extraction from soils: Oil bias from new microbial diversity analysis methods Applied Environmental Microbiology. 67: 2354-2359.
- Allan RN, Lebbe JP, Heyrman P, De Vos CJ, Buchanan, Logan NA. Brevibacilluslevickii sp nov and Aneurinibacillusterranovensis sp nov two novel thermoacidophiles isolated from geothermal soils of Northern Victoria Land, Antarctica. International Journal of Systematic and Evolutionary Microbiology. 2005; 55: 1039-1050.
- Phoebe CH, Combie J, Albert FG, Tran KV, Cabrera J, Guo Y, et al. Extremophilic organisms as an unexplored source for antifungal compounds. Journal of Antibiotics. 2001; 54: 56-65.
- Chaudhuri SR, Pattanayak AK, Thakur AR. Microbial DNA extraction from samples of varied origin” Current Science. 2006; 91: 1697-1700.
- Ekanayake WMJY, Vivehananthan K, Wickramarachchi T. Identification and classification of xenobiotic degrading bacteria in different waste types” Proceedings of the 10th Agricultural Research Symposium, Wayamba University of Sri Lanka. 2010; 65-69.