
Rapid Communication
Austin J Biotechnol Bioeng. 2019; 6(1): 1103.
Transcriptome-Scale Developing of Simple Sequence Repeat Markers in Coffee Arabica
Huang X1, Gbokie T2, Liu BH2, Wu WH1* and Yi KX1*
¹Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, China
²College of Plant Protection, Nanjing Agricultural University, China
*Corresponding author: Wu WH and Yi KX, Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, 4 Xueyuan Road, Haikou 571101, China
Received: April 22, 2019; Accepted: July 19, 2019; Published: July 26, 2019
Abstract
Coffee is an important beverage crop in the world and the main commercially cultivated species are Coffea arabica (arabica) and C. canephora (robusta). C. arabica is a dominant coffee specie with a high potential for genetic improvement and it normally starts fruiting three years after planting, thereby significantly extending its breeding process. Thus, molecular marker assisted selection would efficiently accelerate the process. In the present study, we conducted a largescale development of Simple Sequence Repeat (SSR) markers according to a high quality 454-pyrosequencing database. 1032 SSR loci were identified from 929 unigenes (6.66% of 13951). Mononucleotides (500, 48.45%), trinucleotides (411, 39.83%) and dinucleotides (98, 9.49%) were the main SSR types. The most abundant SSR motif was A/T (490, 47.57%), followed by AAG/CTT (126, 12.23%), ACG/CGT (88, 8.54%), ACT/AGT (70, 6.79%) and AG/CT (61, 5.92%). A total of 115 pairs of reported SSR primers were utilized for the SSR validation and two matched our results. Our work will expand the number of SSR loci and benefit relevant studies by applying these loci in C. arabica.
Keywords: Coffea arabica; Transcriptome; Simple sequence repeat markers
Abbreviations
AFLP: Amplified Fragment Length Polymorphism; SSR: Simple Sequence Repeat; SNPs: Single Nucleotide Polymorphisms; MISA: MIcroSAtellite; COG: Clusters of Orthologous Groups; EST: Expressing Sequence Tag
Introduction
Coffee is an important beverage crop in the world and goblally, the two main species of coffee that are commercially produced are Coffea arabica (arabica) and C. canephora (robusta). C. arabica, the dominant coffee species, contains a high potential for genetic improvement and normally starts fruiting about three years after being planted, which has significantly extended the crop breeding process [1]. Thus, molecular marker assisted selection would efficiently accelerate the process [2]. To date, molecular markers have been successfully utilized in germplasm evaluation of coffee, such as Amplified Fragment Length Polymorphism (AFLP) and Simple Sequence Repeat (SSR) [3,4]. But the markers employed in these studies are still on small amounts. In recent years, the fast development of next generation sequencing technology makes it possible for large scale marker-based germplasm evaluation [5]. A recent study has reported 1444 Single Nucleotide Polymorphisms (SNPs) associated with caffeine content by a draft genome sequence of C. arabica [1]. Although this genome data is still not released with the published study, transcriptome data in previous studies makes it possible for large scale SSR marker developing [6,7]. In this study, we conducted a large-scale development of SSR markers based on a high quality 454-pyrosequencing database [6]. This work has expanded the number of SSR loci which could complement available information and enhance future research efforts on the utilization of these loci in C. arabica.
Materials and Methods
Identification and characterization of SSRs
De novo assembly of 13,951 unigenes of C. arabica CIFC H147/1 from a previous study [6] was utilized for SSR detection by using the MIcroSAtellite (MISA) identification tool with default criteria [8]. The maximal number of bases interrupting two SSR motifs in a compound microsatellite was 20. All the SSR-contained unigenes were searched against the Clusters of Orthologous Groups (COG) data by BLASTx set and then classified into COG categories with a cutoff Expected value (E-value) of 1e–5 [9].
SSR validation with previous study
Primers from previous studies were downloaded to validate the SSRs in the present study [5,10]. These primers were transformed into FASTA format and each primer considered as a single sequence. Then, comparison was conducted by BLASTn-short procedure to search all the unigenes with SSR loci [11]. Each pair of primers that matched the same sequence with a sequence similarity over 95% was selected for SSR loci comparison. If these SSRs were the same with our result, they would be highlighted and also a reliable proof for our work.
Results
Identification of SSRs
A total of 13,951 unigenes from the previous study were utilized for SSR loci screening. As a result, we found 1,032 SSR sites in 929 unigenes with a frequency of 1 SSR per 8.53 kb sequences (Table 1). Among these, 87 unigenes had more than one SSR loci. 79, 6 and 4 sequences contained 2, 3 and 4 SSR loci, respectively. We separately counted the number of unigenes that contained SSR loci and used for SSR detection at different length intervals (Figure 1). 3.3%, 8.0%, 13.6%, 17.1%, 21.4% and 20.2% of unigenes contained SSR loci at 6 different length intervals, respectively.
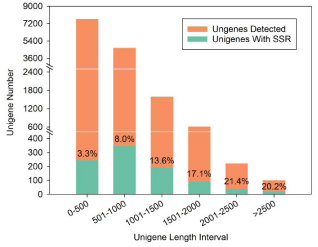
Figure 1: Length distribution of unigenes for SSR detection (orange) and with
SSR loci (green). The percentages in bar represent the ratio of unigenes with
SRR to unigenes for SSR detection.
Item
Count
Total unigenes
13,951
Total size of unigenes (bp)
8,808,436
SSR loci
1,032
Unigenes contain SSR
929
Sequences containing more than 1 SSR
87
Mononucleotide
500
Dinucleotide
98
Trinucleotide
411
Tetranucleotide
15
Pentanucleotide
5
Hexanucleotide
3
Table 1: Results for SSR detection.
Mononucleotides (500, 48.45%), trinucleotides (411, 39.83%) and dinucleotides (98, 9.49%) were the main types of the 1,032 SSR loci (Table 1). Meanwhile, only 15 tetranucleotides, 5 pentanucleotides and 3 hexanucleotides were observed. Among all the identified SSR motifs, the most abundant SSR motif was A/T (490, 47.57%), followed by AAG/CTT (126, 12.23%), ACG/CGT (88, 8.54%), ACT/AGT (70, 6.79%) and AG/CT (61, 5.92%) (Figure 2).
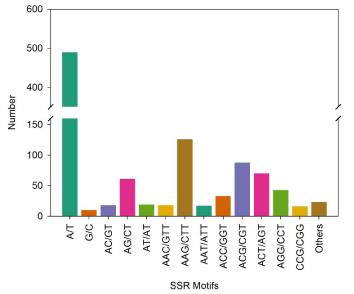
Figure 2: Numbers of SSR motifs.
COG annotation
The 929 unigenes with SSR loci were classified into 25 COG categories (Figure 3). Among these, function unknown (of those poorly characterized and labeled in green, 456) was the most abundant category. And general function prediction only (of those poorly characterized and labeled in green, 62), posttranslational modification, protein turnover, chaperones (of cellular processes and signaling that were labeled in blue, 54), signal transduction mechanisms (of cellular processes and signaling that were labeled in blue, 50) and translation, ribosomal structure and biogenesis (of information storage and processing that were labeled in yellow, 47) were at middle amount.
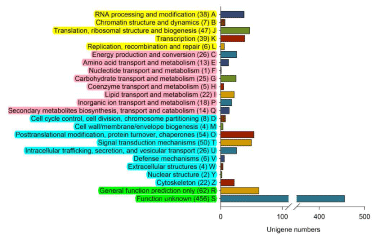
Figure 3: COG classification of the unigenes that containing SSRs and
Arabic numerals represent unigenes numbers. The functional classifications
were listed on the left of y-axis, among which consist of the four functional
categories were labeled in yellow (information storage and processing), pink
(metabolism), blue (cellular processes and signaling) and green (poorly
characterized), respectively.
SSR validation
A total of 115 pairs of published primers were used for SSR validation [3,10]. As a result, two pairs matched unigenes in the current study (Table 2). They shared the same SSR loci but with different sizes of PCR amplification products in CIFC H147/1, respectively.
SSR
Loci
Units
Present Study
Previous Study
Name
Unigenes
Length
Start
Stop
Size
Name
Size
AG
6
Contig20847.505.ag.6
Contig20847
581
505
516
140
SSR03
142,148
AAAGG
3
Contig21585.776.aaagg.3
Contig21585
1160
776
790
135
SSR06
126
Table 2: Validated SSR loci with previous study [10].
Discussion
In plant research works, more SSR markers are necessary for the construction of high-density linkage map and application in breeding processes. In this research, we successfully conducted a large-scale development of SSR markers according to previous tr anscriptome database [6]. The 1,032 Expressed Sequence Tag (EST)-SSR loci have significantly expanded SSRs for C. arabica. Only 6.66% of all the unigenes contained SSR loci, which is relatively lower than other plants, such as 12.29% in kenaf and 7.25% in ramie [12,13]. This observation might have resulted due to the sequence length. The sequence lengths of 11758 unigenes were not over 1000 bp, among which, less than 8% unigenes contained SSR loci. Meanwhile, more than 13% of those unigenes contained SSR loci with sequence lengths over 1000 bp (Figure 1). The result indicated that long unigenes were very possible to contain more SSR loci than short unigenes, which might be also responsibility for rare appearance of tetranucleotides, pentanucleotides and hexanucleotides. Besides, the motifs with high GC region were also infrequent, such as G/C and CCG/CGG. We speculated that a regular successive G/C array might not reach over 30 bp in ESTs of C. arabica [13].
We further classified the COG categories of the 929 unigenes with SSR loci and found that more than half of the unigenes were characterized as function unknown and general function prediction only, which were relatively lower than the previous study in ramie [13]. The poorly annotated unigenes have restricted the functional characterization of these EST-SSR loci. Therefore, we collected a series of reported SSR markers to validated them. Interestingly, our SSR loci didn’t match any of the 103 SSR loci from a previous EST-derived SSR marker development [3]. We just found two (2) pair of SSR markers consistent with our result (Table 2). SSR03 has been reported as a polymorphic SSR loci and SSR06 without polymorphism [10]. However, SSR06 actually has different amplification size in CIFC 147/1, which means it should be considered as a polymorphic SSR loci. In fact, there are still a lot of reported SSR markers that has been utilized for validating our result. More works would be needed in future study, especially PCR amplification. This study has supported a framework and guidance for further study.
Funding
This study was funded by National Key R&D Program of China (2018YFD0201100), FAO/IAEA Collaborative Research Project (20380), International Exchange and Cooperation Project of the Ministry of Agriculture (SYZ2019), Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630042017021, 1630042019030), Special Funds for Efficient Tropical Agriculture Development of Hainan Province (UF37721).
Acknowledgment
We would like to thank Margit Laimer from University of Natural Resources and Life Sciences, BOKU-VIBT (Vienna 1190, Austria) for her thorough suggestions on the experimental design.
References
- Tran HTM, Ramaraj T, Furtado A, Lee LS, Henry RJ. Use of a draft genome of coffee (Coffea arabica) to identify SNPs associated with caffeine content. Plant Biotech J. 2018; 16: 1756-1766.
- Moose SP, Mumm RH. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008; 147: 969-977.
- Poncet V, Hamon P, Minier J, Carasco C, Hamon S, Noirot M. SSR crossamplification and variation within coffee trees (Coffea spp.). Genome. 2004; 47: 1071-1081.
- Gichuru EK, Agwanda CO, Combes MC, Mutitu EW, Ngugi ECK, Bertrand B, et al. Identification of molecular markers linked to a gene conferring resistance to coffee berry disease (Colletotrichum kahawae) in Coffee arabica. Plant Pathol. 2010; 57: 1117-1124.
- Poland JA, Rife TW. Genotyping-by-Sequencing for Plant Breeding and Genetics. Plant Genome. 2012; 5: 92-102.
- Fernandez D, Tisserant E, Talhinhas P, Azinheira H, Vieira A, Petitot AS, et al. 454-pyrosequencing of Coffea arabica, leaves infected by the rust fungus Hemileia vastatrix, reveals in planta-expressed pathogen-secreted proteins and plant functions in a late compatible plant–rust interaction. Mol Plant Pathol. 2011; 13: 17-37.
- Florez JC, Mofatto LS, Freitas-Lopes RL, Ferreira SS, Zambolim EM, Carazzolle MF, et al. High throughput transcriptome analysis of coffee reveals prehaustorial resistance in response to Hemileia vastatrix infection. Plant Mol Biol. 2017; 95: 1-17.
- Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet. 2003; 106: 411–422.
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinform. 2003; 4: 41.
- Geleta M, Herrera I, Bryngelsson T. Genetic diversity of Arabica Coffee (Coffea arabica L.) in Nicaragua as estimated by simple sequence repeat markers. Scientific World J. 2012; 2012: 939820.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinform. 2009; 10: 421.
- Zhang L, Wan X, Xu J, Lin L, Qi J. De novo assembly of kenaf (Hibiscus cannabinus) transcriptome using Illumina sequencing for gene discovery and marker identification. Mol Breeding. 2015; 35: 1-11.
- Chen J,Yu R, Liu L, Wang B, Peng D. Large-scale developing of simple sequence repeat markers and probing its correlation with ramie (Boehmeria nivea L.) fiber quality. Mol Genet Genomics. 2016; 291: 753-761.