
Special Article - Myeloid Leukemia
J Blood Disord. 2015; 2(3): 1032.
DNMT3A Mutations in Tunisian Patients with Acute Myeloid Leukemia
Mechaal Amal¹*, Safra Ines¹, Ben Neji Hind², Rezgui Ichraf¹, Menif Samia¹, Fouzai Chaker¹, Meddeb Balkis² and Abbes Salem¹
1Dpartment of Molecular and Cellular Hematology, University of Tunis El Manar, Tunisia
2Aziza Othmana Hospital, Tunisia
*Corresponding author: Mechaal A, Department of Molecular and Cellular Hematology, Pasteur Institute of Tunis, University of Tunis El Manar, Tunisia
Received: September 14, 2015; Accepted: October 21, 2015; Published: October 28, 2015
Abstract
DNA Methyl Transferase 3A (DNMT3A) is one of two human de novo DNA methyltransferases essential for the regulation of gene expression. DNMT3A mutations were recently linked to hematologic malignancies prognosis. In fact, numerous mutations in this gene were reported in patients with Acute Myeloid Leukaemia (AML), pointing DNMT3A as an important oncogenic role in AML patients. In the present study, 84 patients were analyzed, at the first diagnosis for DNMT3A mutations. Exons 18, 19, 20, 21, 22 and 23 were screened by Polymerase Chain Reaction (PCR) and direct sequencing. The results demonstrated that 33.33% (28/84) de novo AML patients presented DNMT3A mutations. 9 missense mutations including 7 novel single nucleotide polymorphism resulting in amino acid substitution, one silence mutation and 1 nonsense mutation. These mutations are associated with an intermediate-risk cytogenetics (Normal Karyotype (KN-AML)), younger age, higher WBC count, bone marrow infiltration at diagnosis and lower plated count. In conclusion, we retain that the DNMT3A gene is highly mutated in the AML subgroup, its role as a prognostic factor needs to be further elucidated by correlation studies with other molecular prognosis factors and survival.
Keywords: Acute myeloid leukemia; PCR and Direct sequencing; DNMT3A gene
Abbreviations
A: Alanine; AML: Acute Myeloid Leukaemia; bp: Base Pair; C: Cysteine; D: Acide Aspartique; DNA: Desoxyribo Nucleic Acid; DNMT3A: DNA Methyl Transferase 3A; E: Acide Glutamique; F: Phenylalanine; FAB: Franco-Américano-Britannique; g/dl: Grams Per Deciliter; H: Histidine; HSCs: Hematopoietic Stem Cells; I: Isoleucine; K: Lysine; L: Leucine; L: Liter; M: Methionine; N: Asparagines; KN: Normal Karyotype; P: Proline; PCR: Polymerase Chain Reaction; R: Arginine; RNA: Ribo Nucleic Acid; SPSS: Statistical Package for the Social Sciences; V: Valine; WBC : White Cell Count
Introduction
Acute Myeloid Leukemia (AML) is a malignancy of Hematopoietic Stem Cells (HSCs) characterized by the expansion of undifferentiated myeloid progenitors (blasts) with great variability in clinical course and response to therapy. The development of AML is associated with accumulation of acquired genetic alterations and epigenetic changes in hematopoietic progenitor cells that alter normal mechanisms of cell growth, proliferation, and differentiation [1-6]. DNA methylation plays a key role in the pathophysiology of AML [7-10].
DNA Methyltransferase 3A (DNMT3A) is one of two human de novo DNA methyltransferases essential for regulating gene expression during cellular development and differentiation [11]. DNMT3A mutations are generally present in the clones of AML samples, suggesting that they may initiate leukemia [5,6,9,12]. However, the mechanisms by which they contribute to leukemogenesis are not yet clear. Analysis of mutated DNMT3A gene has an important clinical and pathologic application. DNMT3A mutations may become a new tool for target therapy [13, 14].
DNMT3A is located in chromosomal band 2p23 and, is a member of a DNA Methyltransferase (Mtase) family [15]. It has three conserved domains: the PWWP (a highly conserved prolinetryptophan- tryptophan-proline motif) domain targeting the enzyme to nucleic acids, the cystein-rich PHD zinc-finger domain interacting with unmodified histone H3, and the highly conserved catalytic domain representing the Mtase domain in the c-terminal region. DNMT3A has high ubiquitous expression in embryonic tissues and undifferentiated embryonic stem cells. By catalyzing the conversion of cytosine to 5- methylcytosine, DNMT3A add methyl groups to unmodified DNA. DNMT3A mutations, most commonly are heterozygous, and almost at codon R882 in exon 23 DNMT3A Mtase domain modify its enzymatic activity. DNMT3A mutations are significantly enriched in patients with intermediate risk cytogenetics with a normal caryotype [14,16-18].
This work was designed to study the prevalence and nature of the DNMT3A gene mutations in novo AML patients in a Tunisian group.
Patients and Methods
Patients and Bone Marrow Mononuclear Cell (BMMC) collection
We have studied 84 samples of bone marrow from patients with AML, at diagnosis and prior to any chemotherapy. Patients’ samples had been collected from January 2014 to August 2015. Bone marrow mononuclear cells were isolated with Ficoll gradient separation.
The patients’ clinical information’s were obtained from the AML database of different Tunisian hospitals retrospectively. Patients were stratified according to the FAB (Frensh American British) classification.
The diagnosis of the AML subgroup was based on the standard clinic morphologic and phenotype criteria. Cytogenetic information was available for only 78 patients (Table 1).
Characteristics
No DNMT3A
mutation
DNMT3A
mutation
P-value
Patients, n=84
56
28
Age, Years
Median
38.5±9.5
37±19.5
0.041<0.05
Male gender (%)
29 (51.78%)
16 (57.14%)
0.742>0.05
Female gender (%)
27 (48.21%)
12 (42.85%)
Bone marrow blasts at diagnosis (%)
60%
76%
0.015<0.05
WBC count*109/L
Mean
26.6
55.8
0.037<0.05
Hemoglobin, g/dl
Mean
6.95
7.8
0.744>0.05
Platelet count*103/L
Mean
152
60
0.047<0.05
Karyotype, (%)
Normal Karyotype 89%
Abnormal Karyotype 3.57%
Missing Data 7.14%
Normal Karyotype 100 %
0.016<0.05
FAB classification
AML subtype, n = 84
M0
M1
M2
M3
M4
M5
M6
M7
Missing Data
22
1
12
0
12
21
2
2
6
9
0
4
0
3
10
0
0
2
AML: Acute Myeloid Leukaemia; DNMT3A: DNA Methyltransferase 3A; FAB: Franco-Américano-Britannique; KN: Normal Karyotype; g/dl: Grams per Deciliter; L: Liter; WBC: White Cell Count
Table 1: Clinical characteristics of 84 patients with AML.
Genomic DNA isolation, PCR amplification and sequencing
Genomic DNA and total RNA were extracted from mononuclear cells of AML patients using the Trizol method. (Invitrogen Life Technologies- according to the manufacturer’s recommendations), RNA was conserved at -20°C for other potential use. The DNA was extracted following the manufacturer’s instructions.
The extracted DNA was amplified by PCR (S1000 thermal cycler) at the DNMT3A exons 18, 19, 20, 21, 22, and 23, with primers designed by primer 3 tools (Table 2).
Gene
Exon
Sequence 5’-3’
DNA fragment bp
DNMT3A
18-19
TCTCTTTCTTCCTGTCTGCCTCT
GGATGAAGCAGCAGTCCAAG
547
20
TAGAGCAGCACTGTGCAATATG
CTATGGGTCATCCCACCTGC
561
21
TGTGAACTAGTGGCTGCTGG
CACTAGCTGGAGAAGCAGGC
279
22
TAGACGCATGACCAGTGTTGG
TGGAAAACAAGTCAGGTGGG
285
23
TCCTGCTGTGTGGTTAGACG
ATGATGTCCAACCCTTTTCG
417
Table 2: Primer sequences for genes of interest.
PCR was performed in a final volume of 25 ml containing 100 ng of genomic DNA, 2.5 ml of 10X PCR buffer, 1.25 ml of 50 mM MgCl2, 4 pmol of each primer, and 1U of Taq DNA polymerase (Invitrogen, Life Technologies, Carlsbad, CA) The PCR conditions were as follows: denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 62°C for 1 min, extension at 72°C for 1 min. An extension step at 72°C for 10min. The PCR products were sequenced bidirectionally in both the forward and the reverse direction. The same sets of primers used for the PCR amplification were used for sequencing. Samples were sequenced in an ABI PRISM 3130 (Applied Biosystems). Sequencer sequence analysis software version 4.9 (Gene Codes Corporation, Ann Arbor, MI) was used for data analysis.
Statistical analysis
Chi square test was performed by SPSS statistical package version 16.0. Differences were considered significant for when p value lower than 0.05.
Results and Discussion
Molecular analysis of the DNMT3A gene in patients with de novo AML
DNMT3A mutational status was determined in a cohort of 84 de novo AML patients. The sequencing of exons 18, 19, 20, 21, 22 and 23 has revealed 11 mutations. A single-nucleotide polymorphisms L901L that was detected in 3 patients but did not alter the amino acid residues, 7 missense mutations (R736P, R736A (double mutation), P777R, I780M, E784K, M880L and V895M) that were found in 8 patients but had uncertain biologic significance because they were not reported previously and could be verified by a functional study. Two missense mutations were reported previously at exon 23 occurred in 14 patients including the 13 patients with R882H/C. One nonsense mutation (E733) was observed in 3 patients. Most mutations were heterozygous and localized within the catalytic domain. All mutations results were confirmed twice by PCR/sequencing. Sequencing results and description of each mutation are presented in (Figure 1) and (Table 3).
UPNAge/sexKaryotypeFABLocationDNMT3A mutationDNA changeProtein changeP537/246,XXM523c.2645G>Ap.R882HP654/146,XYM219c.2207G>Cp.R736PP731/146,XYM523c.2645G>Ap.R882HP828/146,XYM523c.2644C>Tp.R882CP1124/146,XYM523c.2645G>Ap.R882HP1263/146,XYM019c.2207G>Cp.R736AP1428/146,XYM019c.2197G>TP1557/146,XYM423c.2635 A>Gp.N879DP1647/246,XXM423c.2644C>Tp.R882CP1811/246,XXM023c.2197G>Tp.E733 stop codonP2125/146,XYM023c.2648G>Ap.V895MP2242/246,XXM423c.2645G>Ap.R882HP2833/146,XYM020c.2350G>Ap.E784KP3032/146,XYM023c.2645G>Ap.R882HP3330/146,XYM523c.2644C>Tp.R882CP3763/246,XXM019c.2197G>Tp.E733 stop codonP3837/246,XXM523c.2644C>Tp.R882CP5329/246,XXM523c.2645G>Ap.R882HP5945/146,XYM220c.2350G>Ap.E784KP6449/146,XYMissing Data23c.2638A>Cp.M880LP7221/146,XYM523c.2645G>Ap.R882HP7348/146,XYM523c.2645G>Ap.R882HP7428/246,XXM023c.2703C>Tp.L901LP7539/246,XXM223c.2703C>Tp.L901LP7647/146,XYM523c.2645G>Ap.R882HP8145/246,XXMissing Data23c.2703C>Tp. L901LP8212/246,XXM220c.2330C>Gp.P777RP8349/246,XXM020c.2339T>Gp. I780M
Table 3: Mutation patterns in 28 patients with DNMT3A mutations at diagnosis.
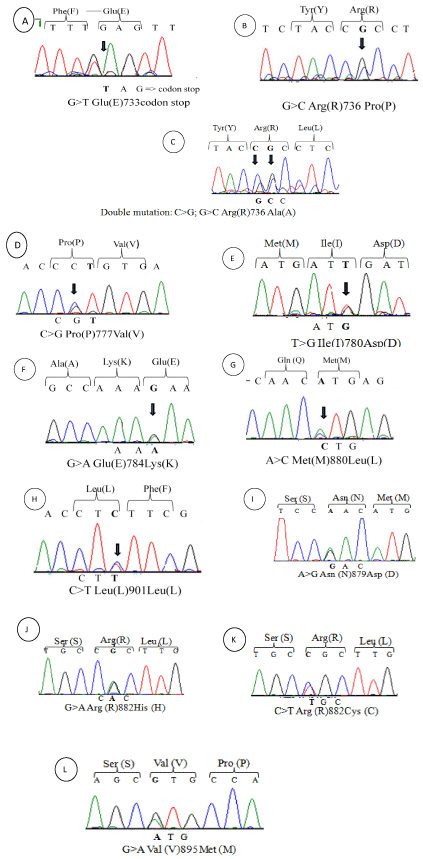
Figure 1: DNMT3A mutations identified in AML: Mutations identified in methyltransferase domain of DNMT3A-Exons 18-23- in 28 patients with AML. Heterozygous
DNMT3A mutations were associated.
(A)E733codon stop (B)R736P/ (C)R736A/ (D)P777V/ (E)I780D/ (F)E784L/ (G)M880L/ (H)L901L/ (I)N879D/ (J)R882H/ (K)R882C/ (L)V895M.
Correlation between the DNMT3A mutations and clinical parameters
The DNMT3A mutation frequency in AML was detected in the heterozygous state in 33.33% (28/84), Compared to no mutated AML group, DNMT3A AML mutation was significantly associated with higher WBC count at diagnosis (p=0.037), bone marrow blasts infiltration (p=0.015), younger adults (p=0.041) and lower platelet count (p=0.047). (Table 1). This high frequency is consistent with results of previous studies on DNMT3A mutations in AML patients which also showed a significant association with higher WBC count, bone marrow blasts, and younger adult age. But this is the first study, in our knowledge that demonstrated a significant association with lower platelet count. [9, 13,19].
The presence of a DNMT3A mutation was found to correlate with AML patients with an intermediate-risk cytogenetics (normal karyotype; CN-AML) (P=0.016), indicating that the mutation of DNMT3A represents a novel prognostic index for intermediate risk AML patients. None of the patients with t (8; 21), or t (15; 17) inv (16) translocation had a DNMT3A mutation (Table 1).
Among 28 patients with AML/DNMT3A mutations, the frequency of DNMT3A R882 mutation was highest (13/28 = 46.42%) which is a mutational hotspot. This is consistent with the studies of Hou et al.; Thol et al. and Yan et al. [13,20,21]. These studies have shown by a quantification of the expression of the cDNA DNMT3A R882 that down expression of DNMT3A may be associated with the incidence and progression of AML.
In our study, AMLDNMT3A R882H/C was significantly associated with myelomonocytic blast morphology (12/13=92.30%, p=1.6*10-5). In fact, 10/10 cases with AML M5 and 2/3 patients with AML M4 were found to harbor DNMT3A R882 mutation , only 1/9 patients with AML M0 was found to harbor DNMT3A R882 mutations and no AMLDNMT3A R882H/C was detected in other’s AML FAB subtypes (Table 1). Moreover in 12/13 AMLDNMT3A R882H/C samples, blasts cells expressed monocytic lineage antigens (CD4, CD14, CD64, CD33) which is in agreement with recent literature [22,23]. This higher significant association could indicate that AMLDNMT3A R882H/C tends to occur in M4 and M5 subtypes in the Frensh American British (FAB) classification. These finding further supported the hypothesis that DNMT3A R882 mutation might be one of the driver mechanism in pre-leukemic stem cell clone.
The analysis of the 7 new mutations R736P, R736A (double mutation), P777R, I780M, E784K, M880L and V895M by software tools polyphen 2 and MutPred [19,24], which predicts whether an amino acid substitution has an impact on the biological function of a protein, demonstrated that these novel mutations, especially R736P, R736A and V895M probably cause protein damage and instability. The assessment of the E733 codon nonsense mutation needs, a functional study would better confirm, or not, this founding for 3 patients.
A number of studies have demonstrated that the activity of DNA methyltransferases may contribute to specific DNA methylation profiles (increased and decreased methylation) and indicated that DNMT3A have a crucial role in the pathogenesis of myeloid acute leukemia. [25-31]. More recently, another interesting study reported no impact of DNMT3A mutation on outcome, but could be a predictive factor for response to idarubicin and thus, could have a direct influence in the way AML patients should be managed [32].
Conclusion and Future Perspective
To our knowledge, this is the first study on the presence of somatic mutations of the gene DNMT3A-exons 18, 19, 20, 21, 22, 23 in patients with de novo AML in Tunisia. Although for a small number of patients 84, we found the frequency of these mutations to be higher comparing with other studies. DNMT3A mutations may represent a new marker for the risk stratification of AML and may have a crucial role in the pathogenesis of myeloid acute leukemia by affecting its expression and methylation of specific genes. Our future aims are quantification of the DNMT3A gene expression, quantification of methylation, evaluation of the new DNMT3A mutations and expanding our study population.
References
- Lowenberg B, Downing JR, Burnett A. Acute myeloid leukemia. N Engl J Med. 1999; 341: 1051-1062.
- Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006; 368: 1894-1907.
- Llopis L, Boissel N, Nibourel O, Wemeau M, Renneville A, Dombret H, et al. Place de la biologie moléculaire dans l’évaluation pronostique des patients atteints de leucémie aiguë myéloïde Molecular biology and prognostic value in acute myeloid leukemia. Hématologie. 2009; 15: 426-443.
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012; 150: 264-278.
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012; 481: 506-510.
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013; 368: 2059-2074.
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009; 361: 1058-1066.
- Alvarez S, Suela J, Valencia A, Fernández A, Wunderlich M, Agirre X, et al. DNA methylation profiles and their relationship with cytogenetic status in adult acute myeloid leukemia. PLoS One. 2010; 5: e12197.
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010; 363: 2424-2433.
- Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrózek K, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012; 30: 742-750.
- Holz-Schietinger C, Matje DM, Harrison MF, Reich NO. Oligomerization of DNMT3A controls the mechanism of de novo DNA methylation. J Biol Chem. 2011; 286: 41479-41488.
- Miller CA, Wilson RK, Ley TJ. Genomic landscapes and clonality of de novo AML. N Engl J Med. 2013; 369: 1472-1473.
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011; 43: 309-315.
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005; 74: 481-514.
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014; 25: 442-454.
- Loghavi S, Zuo Z, Ravandi F, Kantarjian HM, Bueso-Ramos C, Zhang L, et al. Clinical features of de novo acute myeloid leukemia with concurrent DNMT3A, FLT3 and NPM1 mutations. J Hematol Oncol. 2014; 7: 74.
- Pezzi A, Moraes L, Valim V, Amorin B, Melchiades G, Oliveira F, et al. DNMT3A Mutations in Patients with Acute Myeloid Leukemia in South Brazil. Adv Hematol. 2012; 2012: 697691.
- Huang X, Ma D, Dong W, Li P, Lu T, He N, et al. Gene expression profiling of the DNMT3A R882 mutation in acute leukemia. Oncol Lett. 2013; 6: 268-274.
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31: 3812-3814.
- Thol F, Damm F, Lüdeking A, Winschel C, Wagner K, Morgan M, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011; 29: 2889-2896.
- Hou HA, Kuo YY, Liu CY, Chou WC, Lee MC, Chen CY, et al. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012; 119: 559-568.
- Qiao C, Sun C, Zhang SJ, Qian SX, Qian XF, Miao KR, et al. [Analysis of DNMT3a gene mutations in acute myelogenous leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2011; 19: 303-307.
- Marková J, Michková P, Burčková K, Březinová J, Michalová K, Dohnalová A, et al. Prognostic impact of DNMT3A mutations in patients with intermediate cytogenetic risk profile acute myeloid leukemia. Eur J Haematol. 2012; 88: 128-135.
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009; 1: 13.
- Lugthart S, Figueroa ME, Bindels E, Skrabanek L, Valk PJ, Li Y, et al. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood. 2011; 117: 234-241.
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010; 29: 3723-3731.
- Senyuk V, Premanand K, Xu P, Qian Z, Nucifora G. The oncoprotein EVI1 and the DNA methyltransferase Dnmt3 co-operate in binding and de novo methylation of target DNA. PLoS One. 2011; 6: e20793.
- Fathi AT, Abdel-Wahab O. Mutations in epigenetic modifiers in myeloid malignancies and the prospect of novel epigenetic-targeted therapy. Adv Hematol. 2012; 2012: 469592.
- Holz-Schietinger C, Matje DM, Reich NO. Mutations in DNA methyltransferase (DNMT3A) observed in acute myeloid leukemia patients disrupt processive methylation. J Biol Chem. 2012; 287: 30941-30951.
- Jost E, Lin Q, Weidner CI, Wilop S, Hoffmann M, Walenda T, et al. Epimutations mimic genomic mutations of DNMT3A in acute myeloid leukemia. Leukemia. 2014; 28: 1227-1234.
- Gaidzik VI, Schlenk RF, Paschka P, Stölzle A, Späth D, Kuendgen A, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG). Blood. 2013; 121: 4769-4777.
- LaRochelle O, Bertoli S, Vergez F, Sarry JE, Mansat-De Mas V, Dobbelstein S, et al. Do AML patients with DNMT3A exon 23 mutations benefit from idarubicin as compared to daunorubicin? A single center experience. Oncotarget. 2011; 2: 850-861.