
Research Article
Austin J Cancer Clin Res. 2025; 12(1): 1115.
The Role of RANBP3L in Pan-Cancer with its Significance in Hepatocellular Carcinoma
Chao Wang and Jie Li*
Key Laboratory of Integrated Oncology and Intelligent Medicine of Zhejiang Province, Department of Hepatobiliary and Pancreatic Surgery, Affiliated Hangzhou First People’s Hospital, Westlake University School of Medicine, Hangzhou, 310006, China.
*Corresponding author: Jie Li, Key Laboratory of Integrated Oncology and Intelligent Medicine of Zhejiang Province, Department of Hepatobiliary and Pancreatic Surgery, Affiliated Hangzhou First People’s Hospital, Westlake University School of Medicine, No.261, Huansha road, Hangzhou, China Tel: +8615988110485; Fax: +8656005600; Email: 11418184@zju.edu.cn
Received: February 02, 2025; Accepted: February 17, 2025 Published: February 20, 2025
Abstract
Background: RAN binding protein 3-like (RANBP3L), a member of the Ran-binding protein family, has been linked to various cellular functions, but the role in cancer remains underexplored. In this research we assessed the diagnosis and prognosis value about RANBP3L in pan-cancer, especially in liver hepatocellular carcinoma (LIHC).
Methods: We analyzed RANBP3L expression of 33 cancer types from TCGA data and assessed its relationship with overall survival (OS), diseasespecific survival (DSS), and progression-free interval (PFI). We used TIMER2.0 and CIBERSORT to explore the correlation between RANBP3L expression and immune cells. we conducted immunohistochemistry, qRT-PCR, and western blotting by using tissue samples from LIHC patients to assess the RANBP3L’s diagnostic and prognostic value of LIHC.
Results: In 18 different cancers, RANBP3L expression was found to be lower in tumor tissues compared to normal tissues, including LIHC. Lower RANBP3L expression was referred to shorter OS, DSS, and PFI in LIHC. ROC analysis and nomogram model based on RANBP3L expression demonstrated high predictive accuracy for patient diagnosis and survival. Moreover, immune infiltration analysis showed that RANBP3L related to various immune cells and impacted prognosis. Furthermore, analysis on LIHC patient tissues found that higher RANBP3L expression was related to better tumor-free survival and OS.
Conclusion: RANBP3L plays a crucial role in LIHC. Not only does its expression level correlate with patient survival, but it also plays an important role in immune modulation. RANBP3L presents a promising candidate for future therapeutic strategies and biomarker for LIHC.
Keywords: RANBP3L; liver hepatocellular carcinoma; Biomarkers; TCGA
Introduction
Cancer is an important global health challenge, with rising incidence and mortality rates each year [1]. Among those prevalent cancers, liver hepatocellular carcinoma (LIHC) is particularly associated with high mortality rates worldwide [2]. Despite ongoing advancements in cancer diagnosis and treatment, the five-year overall survival (OS) rates for many cancers remain low, highlighting the need for more effective therapeutic strategies [3]. While substantial progress has been made in utilizing cancer biomarkers for diagnosis and prognosis in certain cancers [4], the lack of effective biomarkers for liver and other cancers remains a significant barrier. Given the need for innovative diagnostic and therapeutic methods to improve patient outcomes and offer new avenues for treatment [5].
RAN binding protein 3 like (RANBP3L) is from the family of Ranbinding proteins, which featured the presence of the Ran-binding domain [6]. Ran (Ras-related nuclear protein) is a small GTPase from the Ras superfamily, which could regulate nucleocytoplasmic transport of molecules and control cell cycle progression [7]. It has been reported that RANBP3L could modulate bone morphogenetic protein signaling and the mesenchymal stem cells differentiation [8]. There are little researches about RANBP3L in tumors, only one report shows that the immune cell enrichment score of RANBP3L combined with other seven genes can predict outcomes in endometrial cancer patients [9].
This research was to explore the RANBP3L expression and function in multiple cancers by database, especially in LIHC. Then we assessed the relationship between the RANBP3L expression and immune cells infiltration. Finally we utilized LIHC samples to investigate the RANBP3L expression and prognostic value.
Methods
Database-driven Analysis
The expression of RANBP3L in the 33 cancers (Table 1) were obtained from the Cancer Genome Atlas (TCGA) database (https:// portal.gdc.cancer.gov/). The clinical data were come from the TCGA database.
Kaplan-Meier analysis with log-rank test was used to analyze prognosis included OS (overall survival), DSS (disease-specific survival) and PFI (progression-free interval).
For cancers the RANBP3L expression was linked to the prognosis, the association between clinical characteristic and RANBP3L expression was explored. We used Spearman rank test and Wilcoxon rank-sum test for clinical characteristic analysis. the ROC analysis was conducted applying the “pROC” package. The nomogram model of RANBP3L in LIHC was established according to the RANBP3L expression and the clinical stage to predict the OS. The calibration curve was used to evaluate the predictive accuracy of the nomograms for 1-year, 3-year, and 5-year survival outcomes.
We used TIMER2.0 (Tumor Immune Estimation Resource 2.0, http://timer.cistrome.org/) [10] to explore the correlation between RANBP3L expression and the immune cells infiltration of various tumors in TCGA. Multiple algorithms, including TIMER2.0 and CIBERSORT (https://cibersortx.stanford.edu/) [11], were applied for the immune cell estimation. Additionally, we explored the correlation of immune cell infiltration and OS of RANBP3L expression across different cancer types.
We draw the protein-protein interaction (PPI) network according to the STRING data (https://string-db.org/)., with the interaction threshold of 0.4. The Gene Ontology (GO) analysis containing the biological pathways (BP), the molecular functions (MF) and the cellular components (CC). The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was also performed meanwhile. The cluster Profiler R package and the DESeq R package were used for GESA in this study.
Patients and Samples Collection
Tissue microarrays were constructed using 70 LIHC tissue samples which were got from patients that did hepatectomy at the Hangzhou First People's Hospital. Another 6 paired of HCC tissues and the paracarcinoma tissues used for western blot and RT-qPCR were got from patients who did hepatectomy at Hangzhou First People's Hospital. Ethical Committee approved the research, and each patient signed the informed consent form. This research was carried out following the ethical principles outlined in the Declaration of Helsinki. The diagnosis of HCC was confirmed by pathological examination for every patient. The para-carcinoma tissue was defined as tissue at least 1cm away from the margin of the carcinoma tissue.
Quantitative Real‑time Polymerase Chain Reaction (qRT‑PCR)
The RNA extraction specific experimental steps could refer to this literature [12]. GAPDH was applied as internal standard. The following primer sequences were used: RANBP3L: Forward Primer (5′‑3′), AAATCTGTCATTGCTCAACCCA and Reverse Primer (5′‑3), GCTGCTTCATACAGGGTGTCTT; GAPDH: Forward Primer (5′‑3′), GAACGGGAAGCTCACTGG and Reverse Primer (5′‑3′), GCCTGCTTCACCACCTTCT. The results were calculated by the 2-ΔΔCt method, with each sample being tested in triplicate.
Western Blot (WB)
The extraction of protein and the specific experimental steps could refer to this article [13]. TTC39A Polyclonal antibody (No. 17875- 1-AP) was purchased from Proteintech Group, Inc. GAPDH (No. 60004-1-Ig) which was used as internal standard, was also purchased from Proteintech Group, Inc. Immunodetection was performed by an enhanced chemiluminescence (ECL) detection kit. Then the grayscale values were calculated by the Image J software.
Immunohistochemistry
Immunohistochemistry was performed with the tissue microarrays, make by the 70 pairs of LIHC tissues and the paracarcinoma tissues. The specific experimental steps could refer to this literature [13]. TTC39A Polyclonal antibody (No.21323-1-AP) was purchased from Proteintech Group, Inc. The H-Score was used to estimate the results of immunohistochemistry, using the Visiopharm software. The H-score ranged from 0 to 300, the higher values means stronger overall positive intensity [14].
Results
Comparison of RANBP3L Expression between Normal Tissues and Cancer Tissues
RANBP3L expression for un-paired samples were displayed in 33 cancers according to the TCGA database. Among these 33 cancers, RANBP3L expression was reduced in tumor tissues compared to normal tissues in 18 cancers, including LIHC, KIRC, LUAD and so on (Figure 1A). Similarly, the expression of RANBP3L for paired samples were also found in 23 cancers according to the TCGA database. Among these 23 cancers, RANBP3L expression was reduced in tumor tissues compared to normal tissues in 14 cancers, including LIHC, KIRC, LUAD and so on (Figure 1B).
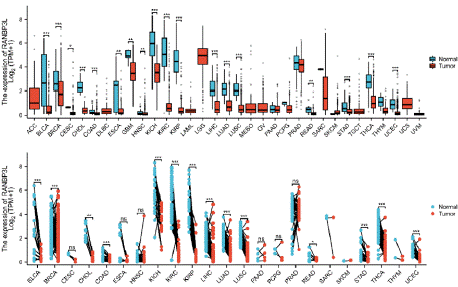
Figure 1: Comparison of RANBP3L expression between cancer tissues
and normal tissues. (A) Comparison of RANBP3L expression between
cancer tissues and normal tissues in un-paired samples. (B) Comparison of
RANBP3L expression between cancer tissues and normal tissues in paired
samples. T test was used, *P<0.05, **P<0.01, ***P<0.001, ns: not significant.
RANBP3L Expression was Correlated with the Patients’ Prognosis
The RANBP3L expression was linked to patients’ OS of with LIHC (Figure 2A). For LIHC patients, the lower expression of RANBP3L in cancer tissues indicated worse OS, respectively (Figure 2B).
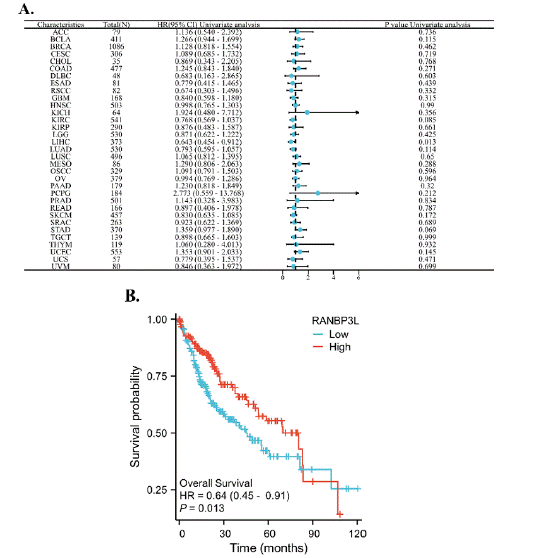
Figure 2: The relationship between the expression of RANBP3L and the OS of patients with 33 cancers, respectively. (A) a forest map, which showed the relationship between the expression of RANBP3L and the OS of patients, for 33 cancers, respectively. (B) the expression of RANBP3L was significantly associated with the OS of patients with LIHC. P<0.05 was considered significant.
The RANBP3L expression was correlated with patients’ DSS with KIRC, LIHC, LUAD, STAD and UCEC (Figure 3A). For KIRC, LIHC and LUAD, lower RANBP3L expression in cancer tissues was associated with poorer DSS (Figure 3B-D). For STAD and UCEC, the higher RANBP3L expression in cancer tissues indicated worse DSS, respectively (Figure 3E, F).
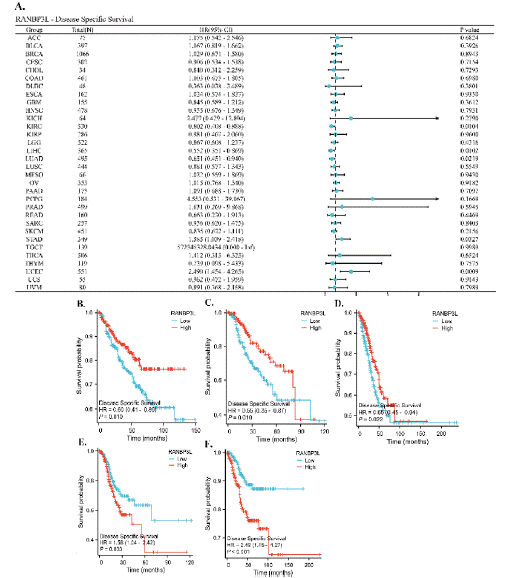
Figure 3: The relationship between the expression of RANBP3L and the DSS of patients with 33 cancers, respectively. (A) a forest map, which showed the
relationship between the expression of RANBP3L and the DSS of patients, for 33 cancers, respectively. (B-F) the expression of RANBP3L was significantly
associated with the DSS of patients with KIRC, LIHC, LUAD, STAD and UCEC, respectively. P<0.05 was considered significant.
The RANBP3L expression was correlated with patients’ PFI with KIRC, STAD and UCEC (Figure 4A). For KIRC, the lower RANBP3L expression in cancer tissues mean worse PFI (Figure 4B). For STAD and UCEC, the higher RANBP3L expression in cancer tissues mean worse PFI (Figure 4C, D).
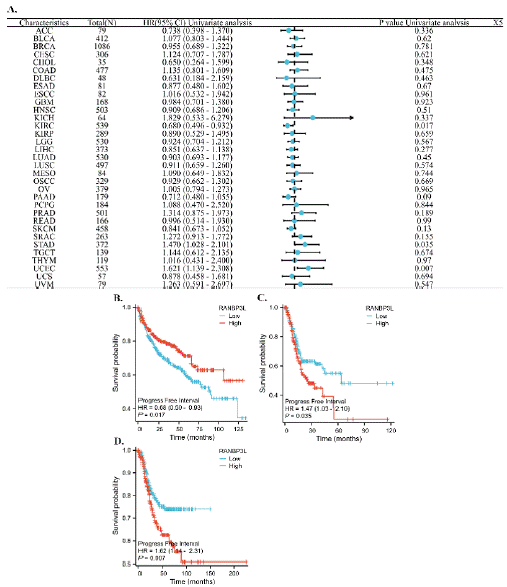
Figure 4: The relationship between the expression of RANBP3L and the PFI of patients with 33 cancers, respectively. (A) a forest map, which showed the
relationship between the expression of RANBP3L and the PFI of patients, for 33 cancers, respectively. (B-D), the expression of RANBP3L was significantly
associated with the PFI of patients with KIRC, STAD and UCEC, respectively. P<0.05 was considered significant.
RANBP3L Expression was Correlated with Patients’ Clinical Features
For cancers that the RANBP3L expression was related to patients’ prognosis, the connection between RANBP3L expression and patients' clinical profiles was further analyzed, according to the TCGA database (Figure 5). In KIRC, the RANBP3L expression was related to pathologic M stage and histologic grade. The RANBP3L expression was significantly related to pathologic M stage, tumor status, AFP (ng/ml) and histologic grade in LIHC. In LUAD, the RANBP3L expression was significantly linked with smoker. In STAD, the RANBP3L expression was significantly related to pathological grade and pathologic T stage. In UCEC, the RANBP3L expression was significantly linked with histological type and clinical stage.
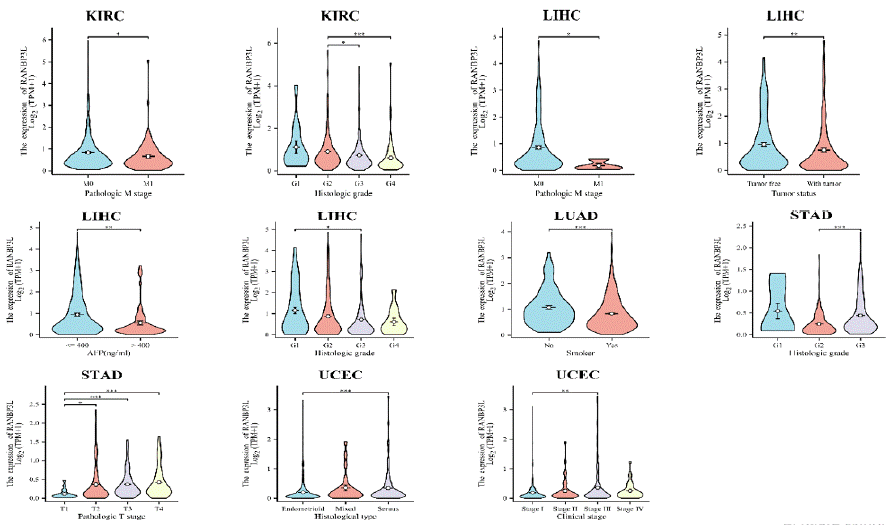
Figure 5: The expression of RANBP3L was associated with some clinical characteristics for patients with KIRC, LIHC, LUAD, STAD and UCEC, respectively.
*P<0.05, **P<0.01, ***P<0.001.
The ROC Curve and Nomogram Model According to the Expression of RANBP3L in LIHC
Among the 33 cancers, the RANBP3L expression was linked to the LIHC patients’ OS. Then, ROC analysis and nomogram model were established from the expression of RANBP3L in LIHC. The ROC analysis showed RANBP3L could be a diagnostic indicator for LIHC, with an Area Under Curve about 0.859 (Figure 6A). The nomogram model demonstrated high precision in estimating the 1-year, 3-year, and 5-year survival probabilities for LIHC patients, as evidenced by the well-calibrated survival prediction curves for these time points. (Figure 6B, C).
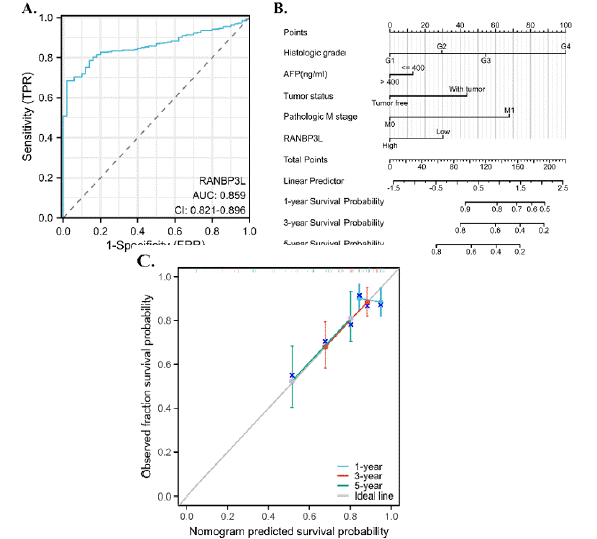
Figure 6: Diagnostic and prognostic effect of RANBP3L in LIHC. (A) RANBP3L could be a good diagnostic indicator for LIHC patients. (B) and (C) RANBP3L
could accurately predict the 1-year, 3-year and 5-year survival probability for LIHC patients.
The RANBP3L Expression was Associated with the Immune Cells Infiltration and could Affect the LIHC Patients’ OS Combined with the Immune Infiltration Score
CIBERSORT analysis showed that in some of cancer types, a relationship was observed between the levels of immune cell infiltration and RANBP3L expression (figure 7). RANBP3L expression had a relation with infiltrating B cells, CD8 T cells, Macrophages, Neutrophils, NK cells, pDC, T cells, Tcm, TFH, Th17 cells, and Th2 cells in LIHC. Neutrophils, macrophage, macrophage/monocyte and common lymphoid progenitor infiltration was related to OS in LIHC.
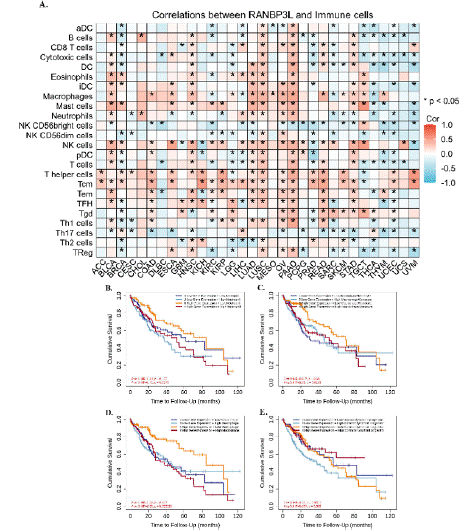
Figure 7: (A) Relationship between RANBP3L expression and immune cell infiltration in different cancers. *p < 0.05. The effect of immune cells infiltration on OS
was related to the expression of RANBP3L. (B–E) Effect of Neutrophils, macrophage, macrophage/monocyte and common lymphoid progenitor infiltration on OS
of LIHC at different RANBP3L expression levels.
The Potential Functions of RANBP3L in LIHC
The top 100 genes associated with the RANBP3L gene were presented as a PPI network in Figure 8A. According to the 100 genes, GO enrichment analysis was conducted including BP analysis, CC analysis and MF analysis (Figure 8B). The BP analysis indicated that RANBP3L might play roles in the process of cellular hormone metabolic, isoprenoid metabolic, terpenoid metabolic and retinoic acid metabolic. The CC analysis indicated that RANBP3L might play roles in microbody and peroxisome. The MF analysis suggested that RANBP3L could be involved in functions such as oxidoreductase activity (utilizing NAD or NADP as acceptors), oxidoreductase activity (targeting CH-OH group of donors), CoA-ligase activity and NAD binding. According to the 100 genes, KEGG pathway analysis was also conducted. The results indicated that RANBP3L might be involved in the process of fatty acid degradation, metabolism of xenobiotics by cytochrome P450, drug metabolism - cytochrome P450 and retinol metabolism(Figure 8C). Furthermore, the potential functions of RANBP3L in LIHC were explored via the GSEA. The result showed that RANBP3L might paly roles in ' Fatty Acid Metabolism ' and ' Biological Oxidations ' (Figure 8D).
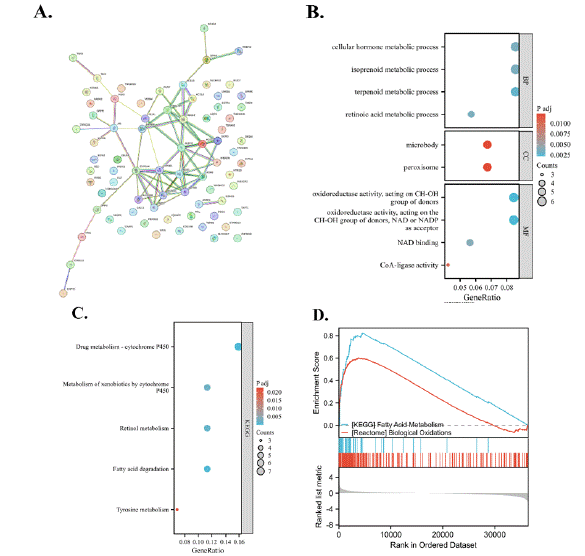
Figure 8: Functional analysis for RANBP3L. (A) PPI network with top-100 genes that related to RANBP3L. (B) Go enrichment analysis. (C) KEGG pathways
analysis. (D) GSEA of RANBP3L in LIHC.
The Expression and Survival of RANBP3L in LIHC Patients
Immunohistochemistry was conducted in 70 LIHC tissues of tissue microarray. We use 235 as the cutoff value of H-Score to distinguish high and low expression of RANBP3L. RANBP3L expression is elevated in para-carcinoma tissues in comparison with LIHC tissues (Figure 9A). Higher RANBP3L expression in LIHC tissues indicated better OS and TFS for LIHC patients (Figure 9B and 9C). Furthermore, the mRNA and protein level of para-carcinoma tissues is higher than LIHC tissues in another 6 LIHC patients (Figure 10).
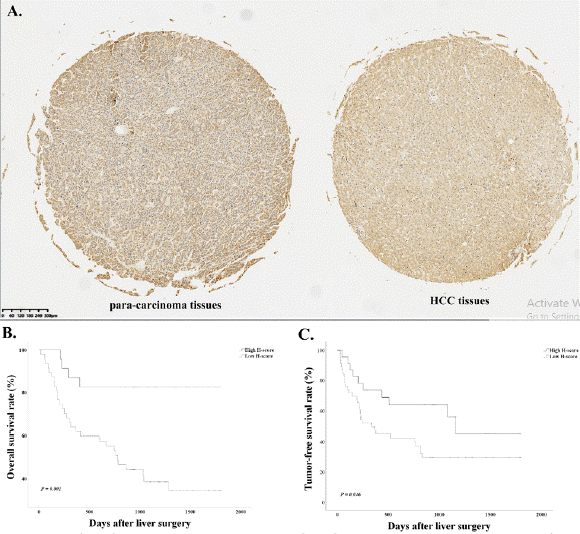
Figure 9: Immunohistochemistry and survival analysis of RANBP3L. (A) Immunohistochemistry analysis of RANBP3L. The expression of para-carcinoma tissue
is higher than LIHC tissue. (B) the overall survival (OS) of LIHC patients. the OS of LIHC patients with high RANBP3L expression is longer than patients with
low expression. (C) the tumor-free survival (TFS) of LIHC patients. the TFS of LIHC patients with high RANBP3L expression is longer than patients with low
expression.
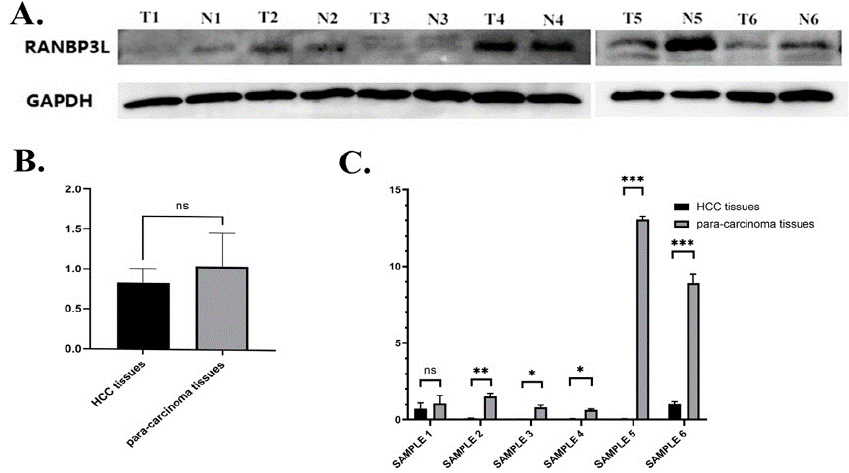
Figure 10: The protein and mRNA expression of RANBP3L in LIHC patients. (A) the protein expression of RANBP3L in LIHC patients, the expression of para-carcinoma tissue is higher than LIHC tissue. (B) Gray value of western blot, the expression of para-carcinoma tissue is higher than LIHC tissue without
significance. (C) the mRNA expression of RANBP3L in LIHC patients. *P<0.05, **P<0.01, ***P<0.001.
Discussion
LIHC is still a leading cancer-related mortality globally, and the development of effective biomarkers for diagnosis and prognosis should be an area of intense research [3]. We analyzed the expression trends and prognostic relevance of RANBP3L across multiple cancers, particularly focus on LIHC to found the role of RANBP3L in the LIHC progress.
We got the RANBP3L is corrected with LIHC clinical characteristics such as pathologic M stage, tumor status, AFP (ng/ ml) and histologic grade. Further research found the foundation of a prognostic model incorporating RANBP3L expression and clinical features demonstrated its predictive accuracy for survival outcomes in LIHC. The nomogram with LIHC clinical characteristics provides a promising tool for individualized prognosis prediction, and this reduction correlates with poor OS and TFS in LIHC patients. we found RANBP3L is linked to important biological pathway such as oxidoreductase activity and attend cytochrome metabolism which may regulated by phosphatidylinositol 3 kinase (PI3K)/Akt, EMT or TGF-β signaling pathways [15]. Furthermore, this study is the first research to explore the RANBP3L protein and mRNA in clinical LIHC samples. The result showed that RANBP3L might function as a candidate tumor suppressor and the function of tumor suppressor may be achieved through regulating BMP-driven lineage-specific differentiation of mesenchymal stem cells [8].
Immunotherapy of LIHC is attracted more and more attention and become very important in the LIHC treatment in recent years [16,17]. In this research, we found the link between RANBP3L and immune cell infiltration. Immune environment signature parameters can assess prognosis in some kind of cancer types [18], including LIHC [19]. These parameters can be used as prognostic elements after immunotherapy [20]. The infiltration of immune cells has been shown to significantly affect LIHC prognosis, assuming its possible regulating LIHC development by the tumor microenvironment remodeling [21].
However, our research has several limitations. Firstly, this study mainly depends on retrospective cohorts and database, and further validation is required. Secondly, the mechanistic pathways about RANBP3L influences immune cell infiltration and LIHC progression still unclear and need further investigation through more basic experiment.
Conclusion
The RANBP3L expression was lower in multiple cancer tissues relative to the normal tissues and significantly associated the LIHC patients’ OS according to the TCGA database. The ROC curve and nomogram model according to the expression of RANBP3L in LIHC could be a good diagnostic indicator. Analysis of LIHC samples revealed that patients exhibiting high RANBP3L expression showed a better prognosis compared to those with low expression. This research found RANBP3L may provide a suspected therapeutic target and predictive biomarker of LIHC.
Declarations
Conflicts of Interest
The authors stated that they had no competing interests.
Authors' contributions
The main manuscript text was written and figures were prepared by Chao Wang. This research was designed and the article was revised by Jie Li. All authors reviewed the manuscript.
Funding
The article received funding from the Basic Public Welfare Research Plan of Zhejiang Province (No. LQLQ24H030009) and the Basic Public Welfare Research Plan of Zhejiang Province (No. LTGY24H160023).
Availability of Data and Materials
The data utilized are publicly accessible and can be found in the TCGA and UCSC databases.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018; 68: 394-424.
- Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer communications (London, England). 2019; 39: 22.
- Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl). 2021; 134: 783-791.
- Wu YL, Tsuboi M, He J, John T, Grohe C, Majem M, et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N Engl J Med. 2020; 383: 1711-1723.
- Jaszek N, Bogdanowicz A, Siwiec J, Starownik R, Kwaśniewski W, Mlak R. Epigenetic Biomarkers as a New Diagnostic Tool in Bladder Cancer-From Early Detection to Prognosis. Journal of clinical medicine. 2024; 13: 7159.
- Mueller L, Cordes VC, Bischoff FR, Ponstingl H. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS letters. 1998; 427: 330-336.
- Boudhraa Z, Carmona E, Provencher D, Mes-Masson AM. Ran GTPase: A Key Player in Tumor Progression and Metastasis. Frontiers in cell and developmental biology. 2020; 8: 345.
- Chen F, Lin X, Xu P, Zhang Z, Chen Y, Wang C, et al. Nuclear Export of Smads by RanBP3L Regulates Bone Morphogenetic Protein Signaling and Mesenchymal Stem Cell Differentiation. Mol Cell Biol. 2015; 35: 1700-1711.
- Gu J, Wang Z, Wang BO, Ma X. ImmuneScore of eight-gene signature predicts prognosis and survival in patients with endometrial cancer. Frontiers in oncology. 2023; 13: 1097015.
- Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020; 48: W509-w514.
- Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods in molecular biology (Clifton, NJ). 2018; 1711: 243-259.
- Li J, Qiu X, Liu Z, Wang J, Wei Q, Zheng S, et al. Tumor Promoting Effect of Long Non-Coding RNA RP11-556E13.1 and its Clinical Significance in Hepatocellular Carcinoma. Clinical laboratory. 2024; 70.
- Li J, Wang J, Liu Z, Guo H, Wei X, Wei Q, et al. Tumor-suppressive role of microfibrillar associated protein 4 and its clinical significance as prognostic factor and diagnostic biomarker in hepatocellular carcinoma. Journal of cancer research and therapeutics. 2022; 18: 1919-1925.
- McCarty KS, Jr., Miller LS, Cox EB, Konrath J, McCarty KS, Sr. Estrogen receptor analyses. Correlation of biochemical and immunohistochemical methods using monoclonal antireceptor antibodies. Archives of pathology & laboratory medicine. 1985; 109: 716-721.
- Chernyakov D, Groß A, Fischer A, Bornkessel N, Schultheiss C, Gerloff D, et al. Loss of RANBP3L leads to transformation of renal epithelial cells towards a renal clear cell carcinoma like phenotype. Journal of experimental & clinical cancer research: CR. 2021; 40: 226.
- Xue R, Zhang Q, Cao Q, Kong R, Xiang X, Liu H, et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature. 2022; 612: 141-147.
- Liu Y, Xun Z, Ma K, Liang S, Li X, Zhou S, et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J Hepatol. 2023; 78: 770-782.
- Yang K, Halima A, Chan TA. Antigen presentation in cancer - mechanisms and clinical implications for immunotherapy. Nature reviews Clinical oncology. 2023; 20: 604-623.
- Sangro B, Sarobe P, Hervás-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021; 18: 525- 543.
- Jiang P, Sun W, Shen N, Huang X, Fu S. Identification of a metabolismrelated gene expression prognostic model in endometrial carcinoma patients. BMC cancer. 2020; 20: 864.
- Bi KW, Wei XG, Qin XX, Li B. BTK Has Potential to Be a Prognostic Factor for Lung Adenocarcinoma and an Indicator for Tumor Microenvironment Remodeling: A Study Based on TCGA Data Mining. Frontiers in oncology. 2020; 10: 424.