
Research Article
Austin J Cancer Clin Res 2015;2(5):1042.
The Relationship of Mir-21, Mir-126 and Mir-205 to P-Glycoprotein, Mrp1 and Lrp/Mvp in Non-Small Cell Lung Cancer
Zizkova V¹†, Skarda J¹,²†, Janikova M¹,²*, Luzna P³, Radova L², Kurfurstova D¹ and Kolar Z¹,²
¹Department of Clinical and Molecular Pathology and Laboratory of Molecular Pathology, Palacky University Olomouc and University Hospital Olomouc, Czech Republic
²Institute of Molecular and Translational Medicine, Palacky University Olomouc and University Hospital Olomouc, Czech Republic
³Department of Histology and Embryology, Palacky University Olomouc and University Hospital Olomouc, Czech Republic †Authors’ contributions: Veronika Zizkova and Jozef Skarda contributed equally to this work
*Corresponding author: Janikova M, Department of Clinical and Molecular Pathology and Laboratory of Molecular Pathology, Institute of Molecular and Translational Medicine, Palacky University Olomouc and University Hospital Olomouc, Hnevotinska 3, 775 15 Olomouc, Czech Republic.
Received: May 20, 2015; Accepted: June 15, 2015;Published: July 29, 2015
Abstract
Protein transporters P-gp, MRP1 and LRP/MVP participate in the emergence of multidrug resistance (MDR) in non-small cell lung cancer (NSCLC). Their expression is post-transcriptionally regulated by microRNAs (miRNAs). Dysregulation of miR-21, miR-126 and miR-205 is often found in NSCLC. The aim of this study was to determine whether the level of miRNAs is associated with expression of the above mentioned proteins involved in MDR and whether they can be used as prognostic and diagnostic markers. We analysed miR-21, miR-126 and miR-205 in various histological subtypes of NSCLC. Their expression was then correlated with clinico-pathological characteristics, such as progression free survival (PFS), overall survival (OS) and different histological subtypes of NSCLC and, with expression of P-gp, MRP1 and LRP/MVP. We found no significant relationship between miR-21 and miR-126 expression and clinico-pathological parameters. However, miR- 205 levels were significantly increased in squamous cell carcinomas (p<10-6) compared with other histological subtypes of NSCLC. Additionally, the level of miR-205 inversely correlated with P-gp expression in NSCLC patients (p=0.03). Results of this study suggest that miR-205 could be used as a diagnostic marker and its downregulation may indicate the emergence of P-gp mediated drug resistance in NSCLC patients.
Keywords: microRNA; NSCLC; P-gp; MRP1; LRP/MVP
Abbreviations
ADC: Adenocarcinoma; EGFR: Epidermal Growth Factor Receptor; FFPE: Formalin-fixed and Paraffin-embedded; HER3: v-erb-b2 Erythroblastic Leukemia Viral Oncogene Homolog 3; LCC: Large Cell Carcinoma; LRP/MVP: Lung Resistance-related Protein/major Vault Protein; MDR: Multidrug Resistance; miRNA: microRNAs; MRP1: Multidrug Resistance-associated Protein 1; NSCLC: Non-small Cell Lung Cancer; OS: Overall Survival; PFS: Progression Free Survival; P-gp: P-glycoprotein; PTEN: Phosphatase and Tensin Homologue; RNAi: RNA Interference; RT-PCR: Reverse Transcription Polymerase Chain Reaction; SCC: Squamous Cell Carcinoma; SCLC: Small Cell Lung Cancer; TMA: Tissue Microarrays; VEGF: Vascular Endothelial Growth Factor; ZEB: Zinc Finger E-box Binding Homeobox
Introduction
A major cause of cancer mortality worldwide is lung cancer and non-small cell lung cancer (NSCLC) represents approximately 80-85% of all cases. Many NSCLC patients do not respond to therapy due to the emergence of multidrug resistance (MDR). For treatment of advanced forms of NSCLC is frequently treated using radiotherapy, chemotherapy or biological therapies. In recent years, RNA interference (RNAi) has also been introduced [1-3]. RNAi is a molecular mechanism of gene silencing which inhibits gene expression at post-transcriptional level. In human it is govern by small (~22 nt) non-coding, endogenous, single-stranded RNAs, called microRNAs (miRNAs). Based on complementarity, these molecules bind to the target mRNA, which is either completely degraded or prevented from translation, without any split [3]. Currently, according to miRBase 21, 2588 mature human miRNAs are known to regulate many protein-coding genes [4-7]. MiRNAs play an important role in many biological processes, such as proliferation, apoptosis, development, differentiation, DNA damage response and other processes. They can also affect carcinogenesis, chemoresistance and radioresistance and they are involved in diverse regulatory pathways [8-12]. For these reasons, miRNAs often dysregulated in tumours could be classified as a class of oncogenes or tumour suppressor genes and, they might be used as markers for monitoring carcinogenesis [8, 13].
The human microRNA-21 gene (hsa-miR-21), located on chromosome 17q23.2, has been shown to be dysregulated in e.g. breast, colon, pancreatic, stomach, prostate, ovarian and lung cancer [14-16]. Overexpression of miR-21 in lung cancer in never-smokers is probably connected with activated epidermal growth factor receptor (EGFR) signalling [17]. miR-21 supports cell proliferation because it inhibits the tumour suppressor gene Phosphatase and tensin homologue (PTEN). This leads to constitutively active signalling through the PI3K/Akt pathway followed by K-Ras signalling which supports survival and proliferation of tumour cells [18-20]. miR-21 also participates in other processes, such as differentiation, cell cycle progression, apoptosis, tumour invasion and DNA-damage repair processes [21-23]. There is also evidence that the Akt2-dependent pathway activated by hypoxia can support tumour resistance via miR- 21 induction. Chemoresistance accompanied by miR-21 upregulation has been found in breast and ovarian carcinomas, pancreatic cancer, prostate cancer and glioblastomas [24-28].
The expression of human microRNA-126 gene (hsa-miR-126), located on chromosome 9q34.3, has been shown to be dysregulated in hepatocarcinomas, breast, colorectal, cervical and lung cancer [29-31,14,15]. miR-126 is the regulator of Vascular endothelial growth factor A (VEGF-A) and therefore, it has an important role in angiogenesis [29,32,33]. It is also involved in proliferation, differentiation and metastasis [31,34,35]. On the other hand, downregulation of miR-126 decreases the cytotoxic effect of gefitinib in adenocarcinomas cell lines. This can lead to the emergence of resistance to gefitinib [36].
The human microRNA-205 gene (hsa-miR-205), located on chromosome 1q32.2, has been shown to be upregulated in head and neck cancer, bladder cancer and in squamous cell carcinomas [37,38]. Likewise miR-21, miR-205 inhibits tumour suppressor gene PTEN and also regulates Zinc finger E-box binding Homeobox 1 and 2 (ZEB1 and ZEB2) regulating tumour invasion [18,39]. miR- 205 regulation of v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (HER3) receptor activating Akt can sensitize breast cancer to treatment using the tyrosine-kinase inhibitors gefitinib and lapatinib [40]. The pro-apoptotic effect of chemotherapeutic agents was also observed in prostate cancer cells with induced expression of miR-205 [41].
The following study on miR-21, miR-126 and miR-205, whose genes are located in regions frequently amplified (hsa-miR-21 and hsa-miR-205) or deleted (hsa-miR-126) in lung cancer was based on the work of Yanaihara et al. [14]. We correlated levels of these miRNAs with clinico-pathological status of NSCLC patients and with expression of known transporter proteins involved in MDR, such as P-glycoprotein (P-gp), Multidrug resistance-associated protein 1 (MRP1) and Lung resistance-related protein/Major vault protein (LRP/MVP). These proteins are able to efflux anti-cancer drugs from the cells which is one of the main mechanisms of MDR [2].
Materials and Methods
Clinical assessment and patients
Formalin-fixed and paraffin-embedded (FFPE) surgical tissue samples of lung cancer from years 1996-2000 were obtained from the archives of the Department of Clinical and Molecular Pathology, Faculty of Medicine and Dentistry, Palacky University and University Hospital, Olomouc. The group of patients comprised of 65 patients with lung cancer after approval by the ethics committee of University Hospital and Faculty of Medicine and Dentistry, Palacky University Olomouc. The cohort consistent of 65 patients: 56 male and 9 female and age range 33 to 78 years. 31 tumours were classified as adenocarcinomas (ADCs), 26 as squamous cell carcinomas (SCCs), 6 as large cell carcinomas (LCCs) and 2 as small cell lung cancer (SCLC) patients. 19 patients were in stage I/II and 34 patients in stage III/IV. For the rest of the patients, the stage was unknown. The characteristics of patients are shown in Table 1. 21 patients had received chemotherapy. 18 of them underwent platinum based chemotherapy regime and the rest of patients were treated with different chemotherapeutics, such as fluorouracil, doxorubicin or taxanes. The clinico-pathological parameters progression free survival (PFS) and overall survival (OS) were monitored over 15-years.
All patients
Sex
Age
Grade
Histological subtype of NSCLC
SCLC
65
female
male
<60
≥60
I/II
III/IV
ADC
SCC
LCC
2
9
56
28
37
19
34
31
26
6
NSCLC – non-small cell lung cancer, SCLC – small cell lung cancer, ADC – adenocarcinoma, SCC – squamous cell carcinoma, LCC – large cell carcinoma
Table 1: Characteristics of patients.
Tissue microarray construction
Tumour tissue microarrays (TMA) were constructed using 65 formalin-fixed and paraffin-embedded primary lung cancer specimens. The tissue areas for sampling were selected based on visual alignment with the corresponding H&E stained section. Two to four tissue cores were taken from from a tumour block and were replaced into a recipient paraffin block with a tissue microarrayer Galileo TMA CK3500 (BioRep, Milan, Italy).
Immunohistochemical staining of P-gp, MRP1 and LRP/MVP
Indirect immunohistochemistry was used. The TMA sections were deparaffinized. Then LRP/MVP antigen was unmasked in citrate buffer (pH 6) and MRP1 in Target Retrieval Solution, High pH (10x) (Dako, Denmark). The sections for detection of P-gp were not pretreated because the monoclonal antibody UIC2 (1864, Immunotech, Marseille, France), used at a dilution of 1:50, targets the extracellular epitope of P-gp protein [42]. The Monoclonal Antibody to MRP1 (human) (MRPm5) (801-012-C250, Alexis, Lausen, Switzerland) at a dilution of 1:25 and Monoclonal Antibody to LRP [MVP] (human) (LMR5) (801-014-C025, Alexis, Lausen, Switzerland) at a dilution 1:20 were used. Visualisation was made by EnVisionTM+ Dual Link (Dako, Denmark). Nuclei were counterstained with haematoxylin. The preparations were observed under an optical microscope and images were captured with a DP71 camera (Olympus, Tokyo, Japan). The expression of P-gp, MRP1 and LRP/MVP were semi quantitatively assessed in stained tissue sections by estimation of the percentage of positive cells as very low (≤ 10%), low (≤ 30%), moderate (≤ 60%) or high (> 60%).
Isolation of total RNA
Total RNA isolation from corresponding tissue cores obtained by tissue microarrayer Galileo TMA CK3500 [43] was performed using the RecoverAll™ Total Nucleic Acid Isolation Kit for FFPE (Applied Biosystems, Foster City, CA, USA) according to the manufacturer´s instructions. All preparation and handling steps of RNA were performed under RNase-free conditions. The concentration of total RNA was measured using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, Delaware, USA) and then RNA was stored at -80°C until use.
Reverse transcription
TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used according to the manufacturer protocol. 10 ng of total RNA was used for reverse transcription in total volume of reaction 15 μl. Pooled gene-specific primers of miR-21, miR-126, miR-205 and RNU6B (RNA, U6 small nuclear 2) (Applied Biosystems, Foster City, CA, USA) were added to the reverse transcription polymerase chain reaction (RT-PCR). RTPCR product was then preamplificated according to protocol using TaqMan® PreAmp Master Mix (2x) (Applied Biosystems, Foster City, CA, USA) with pooled TaqMan® MicroRNA Assays (Applied Biosystems, Foster City, CA, USA) of miR-21, miR-126, miR-205 and RNU6B.
Real-time PCR for miRNAs quantification
The pre-PCR product was diluted in TE buffer (1:20). The reaction consisted of TaqMan® Universal PCR Master Mix (2x) (Applied Biosystems, Foster City, CA, USA), nuclease-free water and corresponding TaqMan® MicroRNA Assay and diluted pre- PCR product. Each sample was analysed in triplicate and volume per each reaction was 10 μl. Real-time PCR was performed using LightCycler® 480 (Roche, Branford, CT, USA) according to protocol. As an endogenous control, RBU6B was used. The control sample was prepared by mixing the same amounts of commercially available Human Lung Total RNAs (AM7968, Lot. 0904002 and 1203010, Applied Biosystems, Foster City, CA, USA) and Total RNAs, Lung, Human (540019, Lot. 0006051745 and 0006079356, Agilent Technologies, Santa Clara, CA, USA).
Statistical analysis
The relative level of expression of assessed miRNAs was calculated using the following equation: relative gene expression = 2-(ΔCt sample – ΔCt control). The difference among controls was determined using ANOVA (analysis of variance) test. For the correlation of expression levels of tested miRNAs and proteins with histological subtypes a Kruskal- Wallis test was used. PFS and OS were determined using Kaplan- Meier analysis. The significance of P-gp, MRP1 and LRP/MVP levels in relation to the higher or lower miRNAs expression in NSCLC were determined by the Mann-Whitney U test. p≤0.05 was considered statistically significant.
Results
TMA blocks from 65 patients were immunohistochemically analysed for P-gp, MRP1 and LRP/MVP expression. The tissues from TMAs were also used for the detection of expression levels of miR-21, miR-126 and miR-205.
Expression of P-gp, MRP1 and LRP/MVP in NSCLC
The expression of P-gp, MRP1 and LRP/MVP was assessed in 62 patients. Three patients were excluded from the analysis; two of them for the diagnosis of small-cell lung carcinoma and one for sampling from the metastasis of ADC into adrenal gland. The P-gp was detected in 39 (62.9%) patients, in 18 (29%) its expression was high (Figure 1a). Expression of MRP1 was found in 48 (77.4%) patients. It was high in 26 (41.9%) patients (Figure 1b). The LRP/MVP was detected in 37 (59.7%) patients; high expression was in 20 (32.3%) patients (Figure 1c). Their expression in different histological subtypes is summarised in Table 2. There was no significant difference in P-gp, MRP1 and LRP/MVP expression between ADC, SCC and LCC specimens. For the next analysis, very low expression of proteins was considered as a negative expression while higher expression levels (low, moderate and high) were considered as a positive expression.
Protein
Intensity
ADC [No. (%)]
SCC [No. (%)]
LCC [No. (%)]
Total [No. (%)]
P-gp
very low
9 (30.0)
12 (46.2)
2 (33.3)
23 (37.1)
low
7 (23.3)
6 (23.1)
3 (50.0)
16 (25.8)
moderate
3 (10.0)
2 (7.7)
0 (0)
5 (8.1)
high
11 (36.7)
6 (23.1)
1 (16.7)
18 (29.0)
Total
30
26
6
62
MRP1
very low
6 (20.0)
7 (26.9)
1 (16.7)
14 (22.6)
low
9 (30.0)
7 (26.9)
1 (16.7)
17 (27.4)
moderate
4 (13.3)
1 (3.8)
0 (0)
5 (8.1)
high
11 (36.7)
11 (42.3)
4 (66.7)
26 (41.9)
Total
30
26
6
62
LRP/MVP
very low
11 (36.7)
13 (50.0)
1 (16.7)
25 (40.3)
low
4 (13.3)
2 (7.7)
3 (50.0)
9 (14.5)
moderate
3 (10.0)
5 (19.2)
0 (0)
8 (12.9)
high
12 (40.0)
6 (23.1)
2 (33.3)
20 (32.3)
Total
30
26
6
62
P-gp: P-glycoprotein; MRP1: Multidrug Resistance-associated Protein 1, LRP/MVP: Lung Resistance-related Protein/major Vault Protein; ADC: Adenocarcinoma; SCC: Squamous Cell Carcinoma; LCC: Large Cell Carcinoma
Table 1: Laboratory values at hospital admission.
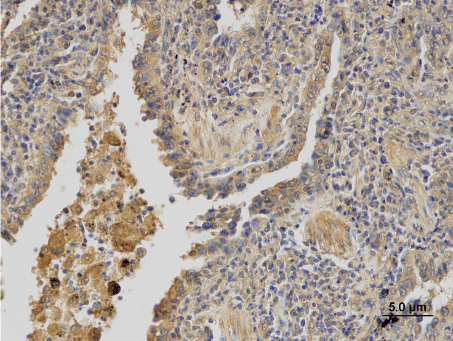
Figure 1a: Immunohistochemical detection of P-gp.

Figure 1b: Immunohistochemical detection of MRP1.

Figure 1c: Immunohistochemical detection of LRP/MVP.
Expression of miR-21, miR-126 and miR-205 in NSCLC
The relative quantification of mature miR-21, miR-126 and miR- 205 in 62 lung cancer samples was performed. Overexpression of miR-21 was found in 7 (11.3%) samples, miR-126 was upregulated in 3 (4.8%) samples and miR-205 was overexpressed in 30 (48.4%) samples. There was no significant difference in miR-21 and miR-126 expression between ADC, SCC and LCC specimens. The expression of miR-205 was significantly higher in SCCs than in other histological subtypes (p<10-6) (Figure 2).
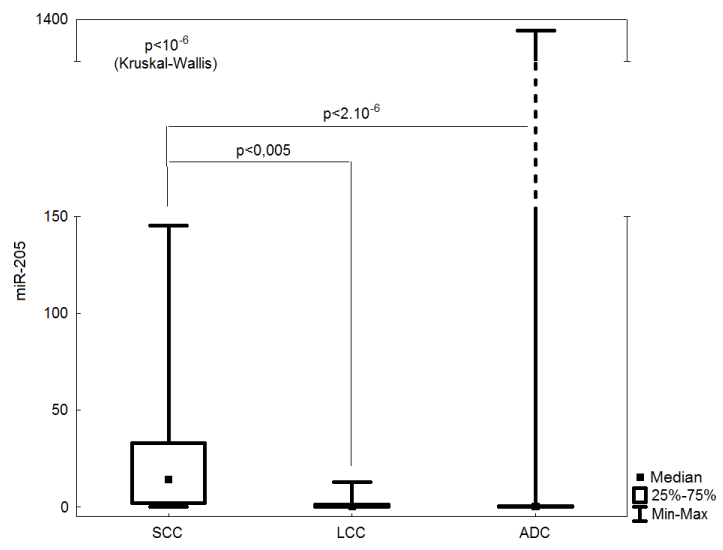
Figure 2: Comparison of miR-205 expression in particular histological
subtypes.
SCC: Squamous Cell Carcinoma; LCC: Large Cell Carcinoma; ADC:
Adenocarcinoma
Analysis of relationship between P-gp, MRP1, LRP/MVP and miRNAs status and, PFS and OS
PFS and OS were used as the clinico-pathological parameters for evaluating the response of patients to the therapy and their survival status. The surveillance analysis did not show the significant differences between the expression of P-gp, MRP1, LRP/MVP and tested miRNAs and, PFS and OS. Expression of these proteins was subsequently correlated with the expression levels of miR-21, mir- 126 and miR-205.
Correlation of miRNAs status with P-gp, MRP1 and LRP/MVP expression
Neither miR-21 nor miR-126 levels correlated with P-gp, MRP1 and LRP/MVP expression. Also the level of miR-205 was not linked with MRP or LRP/MVP expression. The only significant negative correlation was found between miR-205 and P-gp expression (p=0.03) (Figure 3).
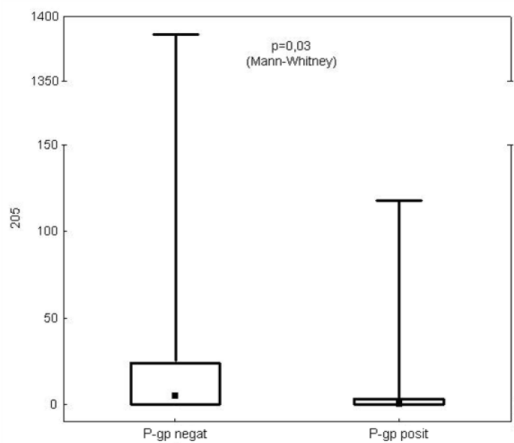
Figure 3: Correlation of miR-205 and P-gp expression.
Discussion
In the present pilot study, data from 62 formalin-fixed and paraffin-embedded primary lung cancer samples were analysed. They were assessed against commercially available control lung RNAs obtained from non-lung cancer patients because the presence of other factors influencing miRNAs expression (e.g. inflammation) in adjacent non-tumour tissues in lung cancer patients.
In this study, no correlation of P-gp, MRP1 and LRP/MVP expression and, miR-21 and miR-126 levels with PFS and OS was found. This is in contrast to other studies in which expression of these proteins and miRNAs appeared to have prognostic value [44-47,13].
The failure of the therapy may be responsible for the shorter PFS and OS of cancer patients probably due to the emergence of MDR. P-gp, MRP1 and LRP/MVP are considered as prognostic markers with regard to the emergence of MDR associated with their expression during chemotherapy [44,45]. Results of this pilot study were not consistent with previous studies. It is probably caused by little cohort of analysed patients because our previous unpublished results obtained from larger group of patients (more than 200) were in agreement with this claim.
In many studies using cell lines, involvement of miR-21 in MDR has been described [24,48-50]. A few studies only have been carried out on NSCLC patient samples. In the majority of publications, miR-21 overexpression is a suggested negative prognostic marker for NSCLC [46,47,13] but in some, this significance was not validated. The downregulation of miR-21 has also been observed and Wang et al. concluded that this may be associated with sensitivity of NSCLC patients to radiotherapy. In this study, the levels of miR-21 in radiosensitive patient’s samples were assessed against radioresistant patient’s samples [51]. There were no available data on the radioresistance / radiosensitivity status of our cohort of patients. However, due to sample collection mainly before therapy, patient radiosensitivity might have been a reason for miR-21 downregulation as downregulation of miR-21 was detected in 55 (88,7%) samples.
Recently, different expression level of miR-21 was observed in distinct tumour areas of colorectal cancer patients. Nielsen et al. found 6-fold higher level of miR-21 in stromal cells compared to that in cancer cells [52]. For the purpose of this study, TMA cores for RNA isolation and quantification were taken from the tumour areas containing cancer cells, not stromal cells. This might explain the finding of lower levels of miR-21 mostly detected in our patients.
Downregulation of miR-126 is commonly observed in NSCLC [34,53] and our results are in accordance with this. In addition, it has been proposed that miR-126 might have a tumour-suppressor effect during tumorigenesis [31].
Our results suggest miR-205 could be used as a diagnostic marker for distinguishing SCCs from ADCs and other histological subtypes (p<10-6). This is consistent with other studies showing the role of miR-205 in NSCLC diagnosis [54-56]. In addition, the relationship of miR-205 and P-gp expression has been observed in this study (p=0.03). So far, there is no verified explanation for this phenomenon but RNA22-HSA algorithm predicted P-gp as one of the targets of miR-205 (https://cm.jefferson.edu/rna22v1.0-homo_sapiens) [57].
P-gp is the main executor of so-called typical MDR. It is responsible for the efflux of some anti-cancer drugs from cells which are one of the major mechanisms of drug resistance. The emergence of the resistance can cause relapse and therefore, its earlier detection might prevent MDR to fully develop. In some cases, typical MDR can be overcome using compounds, such as Cyclosporine A or Tamoxifen [58,59,2]. We present initial results that indicate the possible role of miR-205 in typical MDR caused by P-gp. Validation of these results in a larger cohort of cases is however required.
Conclusion
The recognition of MDR in NSCLC patients is important for decision to change the ineffective chemotherapy to different effective treatment. Detection of molecules regulating expression of proteins involved in MDR may help to reveal these processes earlier. MiR-205 seems to regulate P-gp, which is one of the executors of typical MDR. Therefore, downregulation of miR-205 might be a sign of higher P-gp expression and thus, activation of MDR mechanisms.
Acknowledgement
We would like to thank Jiri Klein and Vitezslav Kolek for kind help with collection and characterisation of patients. We would like to thank also Eva Sedlakova for performance of immunohistochemical staining. This work was supported by grants: IGA MZCR NT13569, LF_2014_003 and NPU I LO1304.
References
- D'Addario G, Früh M, Reck M, Baumann P, Klepetko W, Felip E; ESMO Guidelines Working Group. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010; 21 Suppl 5: v116-119.
- Noskova V, Hajduch M, Mihal V, Cwiertka K. Mechanizmy mnohocetné lékové rezistence a jejich význam pro klinickou praxi, I. typická MDR. Klin Onkol. 2010; 13: 4-9.
- Paranjape T, Choi J, Weidhaas JB. MicroRNA as Potential Diagnostics and Therapeutics. In: MicroRNAs in Development and Cancer. Slack FJ, editor. Imperial College Press. 2011; 213-236.
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004; 32 (Database issue): D109-111.
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006; 34: D140-144.
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008; 36 (Database issue): D154-158.
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011; 39: D152-157.
- Ventura A, Kumar MS, Jacks T. Roles of microRNAs in cancer and development. In: MicroRNAs: From Basic Science to Disease Biology. Appasani K, editor. Cambridge University Press. 2008; 322-337.
- O'Connell RM, Baltimore D. MicroRNAs and hematopoietic cell development. Curr Top Dev Biol. 2012; 99: 145-174.
- Mansfield JH, McGlinn E. Evolution, expression, and developmental function of Hox-embedded miRNAs. Curr Top Dev Biol. 2012; 99: 31-57.
- Wan G, Mathur R, Hu X, Zhang X, Lu X. miRNA response to DNA damage. Trends Biochem Sci. 2011; 36: 478-484.
- Ueno K, Hirata H, Hinoda Y, Dahiya R. Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int J Cancer. 2013; 132: 1731-1740.
- Wang Q, Wang S, Wang H, Li P, Ma Z. MicroRNAs: novel biomarkers for lung cancer diagnosis, prediction and treatment. Exp Biol Med (Maywood). 2012; 237: 227-235.
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006; 9: 189-198.
- Koturbash I, Zemp FJ, Pogribny I, Kovalchuk O. Small molecules with big effects: the role of the microRNAome in cancer and carcinogenesis. Mutat Res. 2011; 722: 94-105.
- Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010; 13: 57-66.
- Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009; 106: 12085-12090.
- Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta. 2010; 411: 846-852.
- Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010; 18: 282-293.
- Frezzetti D, De Menna M, Zoppoli P, Guerra C, Ferraro A, Bello AM, et al. Upregulation of miR-21 by Ras in vivo and its role in tumor growth. Oncogene. 2011; 30: 275-286.
- Gao W, Xu J, Liu L, Shen H, Zeng H, Shu Y. A systematic-analysis of predicted miR-21 targets identifies a signature for lung cancer. Biomed Pharmacother. 2012; 66: 21-28.
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008; 28: 5369-5380.
- Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc Natl Acad Sci U S A. 2010; 107: 21098-21103.
- Polytarchou C, Iliopoulos D, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, et al. Akt2 regulates all Akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res. 2011; 71: 4720-4731.
- Mei M, Ren Y, Zhou X, Yuan XB, Han L, Wang GX, et al. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat. 2010; 9: 77-86.
- Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One. 2010; 5: e10630.
- Shi GH, Ye DW, Yao XD, Zhang SL, Dai B, Zhang HL, et al. Involvement of microRNA-21 in mediating chemo-resistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol Sin. 2010; 31: 867-873.
- Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 2010; 1352: 255-264.
- Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008; 15: 261-271.
- Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008; 10: 203.
- Feng R, Chen X, Yu Y, Su L, Yu B, Li J, et al. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010; 298: 50-63.
- Heusschen R, van Gink M, Griffioen AW, Thijssen VL. MicroRNAs in the tumor endothelium: novel controls on the angioregulatory switchboard. Biochim Biophys Acta. 2010; 1805: 87-96.
- Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, et al. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006; 355: 2542-2550.
- Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, et al. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008; 373: 607-612.
- Huang F, Fang ZF, Hu XQ, Tang L, Zhou SH, Huang JP, et al. Overexpression of miR-126 promotes the differentiation of mesenchymal stem cells toward endothelial cells via activation of PI3K/Akt and MAPK/ERK pathways and release of paracrine factors. Biol Chem. 2013; 394: 1223-1233.
- Zhong M, Ma X, Sun C, Chen L. MicroRNAs reduce tumor growth and contribute to enhance cytotoxicity induced by gefitinib in non-small cell lung cancer. Chem Biol Interact. 2010; 184: 431-438.
- Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005; 33: 5394-5403.
- Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007; 25: 387-392.
- Greene SB, Gunaratne PH, Hammond SM, Rosen JM. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci. 2010; 123: 606-618.
- Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, et al. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009; 69: 2195-2200.
- Bhatnagar N, Li X, Padi SK, Zhang Q, Tang MS, Guo B, et al. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010; 1: e105.
- Kennedy BG, Mangini NJ. P-glycoprotein expression in human retinal pigment epithelium. Mol Vis. 2002; 8: 422-430.
- Cardano M, Diaferia GR, Falavigna M, Spinelli CC, Sessa F, DeBlasio P, et al. Cell and tissue microarray technologies for protein and nucleic acid expression profiling. J Histochem Cytochem. 2013; 61: 116-124.
- Trussardi-Regnier A, Millot JM, Gorisse MC, Delvincourt C, Prevost A. Detection of drug-resistance genes using single bronchoscopy biopsy specimens. Oncol Rep. 2007; 18: 703-708.
- Oue T, Yoneda A, Uehara S, Yamanaka H, Fukuzawa M. Increased expression of multidrug resistance-associated genes after chemotherapy in pediatric solid malignancies. J Pediatr Surg. 2009; 44: 377-380.
- Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008; 54: 1696-1704.
- Gao W, Shen H, Liu L, Xu J, Xu J, Shu Y, et al. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011; 137: 557-566.
- Gao W, Lu X, Liu L, Xu J, Feng D, Shu Y, et al. MiRNA-21: a biomarker predictive for platinum-based adjuvant chemotherapy response in patients with non-small cell lung cancer. Cancer Biol Ther. 2012; 13: 330-340.
- Blower PE, Chung JH, Verducci JS, Lin S, Park JK, Dai Z, et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol Cancer Ther. 2008; 7: 1-9.
- Rui W, Bing F, Hai-Zhu S, Wei D, Long-Bang C. Identification of microRNA profiles in docetaxel-resistant human non-small cell lung carcinoma cells (SPC-A1). J Cell Mol Med. 2010; 14: 206-214.
- Wang XC, Du LQ, Tian LL, Wu HL, Jiang XY, Zhang H, et al. Expression and function of miRNA in postoperative radiotherapy sensitive and resistant patients of non-small cell lung cancer. Lung Cancer. 2011; 72: 92-99.
- Nielsen BS, Jørgensen S, Fog JU, Søkilde R, Christensen IJ, Hansen U, et al. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011; 28: 27-38.
- Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L, et al. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun. 2010; 391: 1483-1489.
- Lebanony D, Benjamin H, Gilad S, Ezagouri M, Dov A, Ashkenazi K, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009; 27: 2030-2037.
- Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010; 16: 610-619.
- Zhang YK, Zhu WY, He JY, Chen DD, Huang YY, Le HB, et al. miRNAs expression profiling to distinguish lung squamous-cell carcinoma from adenocarcinoma subtypes. J Cancer Res Clin Oncol. 2012; 138: 1641-1650.
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006; 126: 1203-1217.
- Claudio JA, Emerman JT. The effects of cyclosporin A, tamoxifen, and medroxyprogesterone acetate on the enhancement of adriamycin cytotoxicity in primary cultures of human breast epithelial cells. Breast Cancer Res Treat. 1996; 41: 111-122.
- Urasaki Y, Ueda T, Nakamura T. Circumvention of daunorubicin resistance by a new tamoxifen derivative, toremifene, in multidrug-resistant cell line. Jpn J Cancer Res. 1994; 85: 659-664.