
Research Article
Austin J Cancer Clin Res. 2019; 6(1): 1084.
Curcumin Nanocrystallites are an Ideal Nanoplatform for Cancer Chemotherapy
Wang X1,2, Peng Y2, Tan H2, Li W2* and Li M1*
¹Zhongshan School of Medicine, Sun Yat-Sen University, China
²Department of Neurosurgery, The First Affiliated Hospital of Shenzhen University, China
*Corresponding author: Mingtao Li, Zhongshan School of Medicine, Sun Yat-Sen University, Guangzhou 510080, China
Weiping Li, Department of Neurosurgery, Shenzhen Second People’s Hospital, The First Affiliated Hospital of Shenzhen University, Shenzhen 518039, China
Received: September 10, 2019; Accepted: October 24, 2019; Published: October 31, 2019
Abstract
Curcumin, a principal active component extracted from rhi-zomes of turmeric, has a promising clinical activity against a wide variety of tumors. It is extremely low solubility in water is the great limitation in the application of cancer chemotherapy. Nanocrystals have recently emerged as a promising pharmaceutical preparations way for the poorly water soluble drugs. Accordingly, we prepared Curcumin Nanocrystals (CNs) with a mean diameter of 15 nm by quick emulsion freeze-drying method. CNs proved to be effective in drug delivery, such as avoiding uptake by the Reticuloendothelial System (RES), prolonging the circulation times of curcumin as well as Enhancing the well-known Permeation and Retention (EPR) effect, as a result of which CNs accumulate at the tumor site. Furthermore, we design to conjugate a targeting ligand-cyclic iRGD peptide to CNs in order to selectively deliver Curcumin to cancer cells with greater efficiency and enhance the permeability of cells leading more drugs inside cells. Hence, our results suggested that iRGD-CN, which be of both characteristics of passive and active targeting, may be a potential drug delivery system in enhance the effective therapy of curcumin chemotherapy.
Introduction
In the preparation of CNs, Pluronic®F-127 were used as emulsion stabilizer and eventually a small amount of them wrapped in the surface of the CNs [1,2]. Pluronic®F-127 are nonionic triblock copolymers composed of a central hydrophobic Chain of poly (Propylene Oxide) (PPO) flanked by two hydrophilic chains of Poly (Ethylene Oxide) (PEO) (PEO-PPO-PEO) [3,4]. These amphiphilic triblock copolymers strengthen stealth properties to CNs, allowing them to reduce potential toxicity and avoid uptake by the RES, which is crucial for achieving long circulation time in blood and enhanced uptake by tumors via EPR effect [5-7]. In addition, the thin coating of Pluronic®F-127 also provide a binding site for cyclic iRGD peptide, which is active targeting ligand enhancing the therapeutic efficacy. iRGD (CRGDK/RGPD/EC) is a kind of tumor-penetrating peptide which can contribute CNs to increasing cancer vascular and tissue permeability in a αv integrin and neuropilin-1 (expressing at the surface of tumor blood vessel endothelial cells and tumor cells)– dependent manner. iRGD-based targeting mechanism for tumor is that the iRGD motif mediates binding to integrins on tumor endothelium and a proteolytic cleavage then exposes a binding motif for neuropilin-1, which mediates penetration into tumor tissue and cells. Compared with traditional targeting moiety simply paying attention to the ability to identify tumor cells and lacking capacity of leading drugs penetration into tumor tissue and cells, iRGD peptide overcomes this limitation and establishes a capability for tissue/ cells-penetrating drug delivery [1,8]. Current research shows that the microenvironment in solid tumors is very distinct from that in normal tissues. Due to deregulated cancer cell metabolism, highly heterogeneous vasculature and defective blood perfusion, the tumor microenvironment is characterized high interstitial pressure and cell density [9,10]. How to make CNs penetrate tumor blood vessel endothelial cells and tumor tissue is the key to eliminate cancer cells, especially to extirpate interior cancer stem cells [11,12]. Thus, we design an efficient DDS through employing the CNs to be chemically conjugated to the peptide.
Materials and Methods
Materials
Pluronic®F-127, 4-(Dimethylamino) Pyridine (DMAP), triethylamine, and succinic anhydride were purchased from Sigmae- Aldrich (St. Louis, MO, USA). Uppsala 87 Malignant Glioma (U87) were supplied by American Type Culture Collection (ATCC). Dulbecco’s Modified Eagle’s Medium (DMEM), RMPI 1640 medium, Fetal Bovine Serum (FBS) were provided from Gibco BRL (Grand Island, NY, USA). Curcumin, Cremophor EL, Cell Counting Kit-8 (CCK-8), Rhodamine-phalloidin, and Dimethyl Sulfoxide (DMSO) were purchased from Sigmae-Aldrich (St. Louis, MO, USA). Oregon Green®488 Taxol, Flutax-2, DAPI (4,6-diamidino-2-phenylindole), Hoechst, LIVE/DEAD Viability/Cytotoxicity Kit, and Cy5 was bought from Molecular Probes (Eugene, OR, USA). Taxol (TA) was supplied by Merck (Germany). DSPE-PEG-iRGD were supplied by GL Biochem Ltd. (Shanghai, China). Anhydrous ethanol and dichloromethane were purchased from Beijing Chemical Reagent Company (Beijing, China).
Pluronic®F-127 functional group modification
Terminal hydroxyl groups on Pluronic®F-127 were converted to carboxyl groups in order to efficiently coupling of iRGD peptide to the CNs surface. The modification of Pluronic®F-127 functional group involves the following procedure. Pluronic®F-127 (0.2g) was dissolved in THF (10mL), and then DMAP (12mg), triethylamine (100 μL) and succinic anhydride (600mg) were added. The reaction mixture was stirred for 48 h at room temperature. The solution was dried by a rotary evaporator and was then dissolved in of chloroform (75mL). The excess succinic anhydride was removed by filtration with a 0.45 μm PTFE membrane syringe filter. The Pluronic®F-127-COOH (F127) was purified by precipitation with ice-cold diethylether [13,14]. The product was identified by 1H-NMR spectrometer (Bruker AVANCE III 600MHz) and Fourier Transform Infrared Spectrophotometer (JASCO FT/IR-660).
Preparation and characterization of CNs and iRGD-CNs
CNs were prepared via oil-in-water emulsions freeze-dried method, which generally comprised three steps process. The first step is emulsion preparation. The Oil phase (O) was dichloromethane solution containing 40mg/mL curcumin. Containing 1 wt% Pluronic®F-127-COOH aqueous solution was the Water phase (W). The volume ratio of oil phase and water phase was fixed at 1:20, and the mixture was mixed through ultrasonic (100w, 10 min) emulsification to obtain emulsion (O/W). Secondly, the above emulsion was further added into 20-fold the volume of the water phase, and the mixture was frozen by liquid nitrogen followed by freeze-drying using a vacuum freeze-dryer. Finally, CNs suspension was obtained when lyophilized products were dispersed into the water and free surfactant were removed by ultracentrifugation (800000rcf, 10min). In this study, CNs were functionalized modifications targeted by DSPE-PEG-iRGD ligand were embedded to the surface of CNs during the process of oil-in-water emulsions freeze-dried. In addition, we used fluorescent dye Cy5 instead of curcumin in the preparation progress to obtain nanocrystals with fluorescence properties so as to study the cellular uptake of CNs and the target of iRGD.
Characterization of CNs
Emulsion droplet size and the desired degree of droplet size uniformity play a key role in the preparation of CNs. Hence, we measured the average particle size and size Polydispersity Index (PDI) of droplets using Zetasizer (Nanoseries, Malvern, UK.). CNs size and morphology were determined by Transmission Electron Microscope (TEM, JEOL, Japan) with an acceleration voltage of 200Kv. The homogeneity of the particles was studied by Atomic Force Microscope (AFM, Bioscope Catalyst). Due to the obvious difference zeta potential of the product during each step of the reaction, the reaction process can be easily detected according to the potential variation.
To assess the encapsulation efficiency of curcumin, free curcumin in the CNs suspension was removed by the method of ultrafiltration, and then curcumin containing in CNs was quantified by HPLC.
In Vitro Cellular uptake
U87 cells were routinely cultured respectively in complete DMEM and RMPI 1640 medium with 10% FBS, 1% penicillin and 1 % streptomycin in a humidified incubator with 5% CO2 at 37oC. For the cellular uptake study, U87 cells were seeded at the density of 1×105 cells/well in two 24-well cell plates and incubated at 37oC for 24h to allow cell attachment. After 24h, the cells were then incubated with CNs and iRGD-CNs (100 μg/mL) at 37oC for different time intervals. The cells were rinsed with PBS (pH 7.2) solution softly, pre detached by 0.25% trypsin. The trypsin was removed by centrifugation at 1000rpm for 5min. The cells were then harvested, re-suspended in 500ml PBS and examined by flow cytometry using a CyAn ADP 9 color flow cytometer (Beckman Coulter). Using the same method, U87 cells were grown in a culture dish at a density of 1×106 cells/dish and incubated at 37oC for 24h. Following incubation of cells in dishes culture containing 200 μL of CNs and iRGD-CNs. The medium was then removed and cells were washed with PBS (pH 7.2). Finally, the total fluorescence images of nanocrystals internalized into cells were scanned using an In Vivo imaging system (FX Pro, Kodak) with an excitation band pass filter at 480nm and an emission at 633nm. In the Confocal Laser Scanning Microscopy (CLSM) imaging studies, U87 cells were first seeded on Petri dishes for 24h, and the medium was replaced with new medium containing 100 μg/mL of CNs and iRGD-CNs. There are some differences in the following process of the two cell lines. After incubation for 24h at 37oC, the medium of U87 cells was then removed and cells were washed with PBS (pH 7.2) followed by fixing with 4% paraformaldehyde for 30 min. The cell membrane and nuclei were stained with Rhodamine-phalloidin and DAPI, respectively. CNs and iRGD-CNs were detected fluorescence at 633nm, and the corresponding fluorescent images of the cell membrane and nuclei at 488nm and 405nm by CLSM (TCS SP5, Leica). However, the cell membrane and nuclei of U87 cells were stained as soon as the medium was removed and cells were washed with PBS. The fluorescent dye was Oregon Green®488 Taxol and Hoechst, whose excitation wavelengths were 488nm and 405nm respectively. And then the fluorescent images of the cells were analyzed using a TCS SP2 confocal microscope.
In Vitro ctotoxicity
The In Vitro cytotoxicity of curcumin, CNs, and iRGD-CNs were evaluated by CCK-8 assay. U87 cells were seeded in 96-well plates at the density of 5×105 cells/well and cultured for 24h at 37oC. The medium in each well was refreshed with 200 μL medium containing serial dilutions of curcumin, CNs, and iRGD-CNs (corresponding curcumin concentration was 0-25 μg/mL). After incubation for 48 h, 20 μL of Cell Counting Kit-8 was added to all wells, and then the cells were continued to be incubated at 37oC with 5% CO2 for 4h. Infinite M200 microplate spectrophotometer (Tecan, Mannedorf, Switzerland) was used to detect the absorbance at 450nm. Percent viability was normalized to cell viability in the absence of the samples.
Results and Discussion
Physicochemical characterization of iRGD-CNs
In an attempt to solve poor drug dissolution problem, we prepared CNs by emulsion technique as an effectual strategy to enhance the saturation solubility and dissolution rate of hydrophobic drugs. During the emulsion preparation, we used the hydrophilic material F127 to stabilize the oil-water interface in virtue of its block copolymer structure with amphiphilic property. The size and morphology of the nanoparticles were characterized by Transmission Electronic Microscopy (TEM). The synthesized CNs dispersed well in cyclohexane and had a diameter of about 15nm. This surface structure characteristic endued CNs with excellent dispersion stability in aqueous solution. To integrate tumor-targeted function, DSPE-PEGiRGD ligand was added during the preparation process of CNs as previously described to obtain iRGD -NDs, which presented uniform structure with an average size of around 15nm by TEM image (Figure 1). Dynamic Light Scattering (DLS) analysis exhibited iRGDCNs with a hydrated particle size of 18nm iRGD was successfully conjugated on the surface of CNs. The slightly increased particle size and zeta potential also indicated the success of the iRGD anchoring on the CNs surface (Table 1).
Groups
Particle size (nm)
Zeta potential (mV)
CNs
15±1.0
-6.4±0.57
iRGD-CNs
18±1.2
1.5±0.48
Table 1: Particle size and zeta potential of the CNs and iRGD-CNs agents.
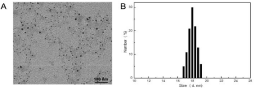
Figure 1: (A) TEM images clearly exhibit the morphologies of iRGD-CNs. (B) Dynamic Light Scattering (DLS) measurement of the as-prepared iRGD-CNs.
To achieve efficient accumulation in the tumor site, these nanoparticles should also possess high binding affinity toward cancer cells. We therefore evaluate the iRGD-conjugated effect on cancer cell uptake. As shown in (Figure 2), only a very small amount of CNs could be internalized into U87 cells due to the charge repulsion between the nanoparticles and the cell membrane, both negatively charged. Once iRGD was conjugated, these nanoparticles could be more extensively internalized. This is because the iRGD could bind preferentially to the integrin receptor that is highly expressed on the surface of U87 cells. This iRGD guided delivery efficiency was further verified by CLSM imaging. These results together reflected that the cellular uptake of iRGD modification nanoparticles was significantly promoted for cancer cells.
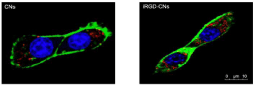
Figure 2: Confocal Laser Scanning Microscopy (CLSM) images of internalization by U87 cells (red spots indicate nanocrystals vesicles, blue indicates cell nucleus,
and green indicates cell membrane).
To test combination treatment effect, we assessed the cytotoxicity of PBS, free curcumin, CNs, and iRGD-CNs with light using CCK- 8 (cell counting kit-8) assay. As shown in (Figure 3), free curcumin showed severe cytotoxicity to U87 cells due to the traditional chemotherapy. As expected, CNs and iRGD-CNs exhibited excellent anti-tumor activities by combining these two methods of treatment for cancer cells. Notably, iRGD-CNs showed a significant improvement in antitumor therapeutic efficacy because of iRGD targeted modification. The corresponding IC50 (concentration resulting in a 50% inhibition of cell growth) values for free curcumin and CNs were more than 10 μg/mL, while that of iRGD-CNs was only about 2 μg/mL.
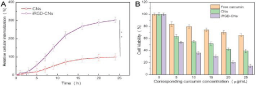
Figure 3: (A) Internalization kinetics of different types of CNs and iRGD-CNs in U87 cells (**p < 0.01). (B) Cytotoxicity of different curcumin formulations against
U87 cells after 48h incubation.
Conclusion
In summary, nanocrystallites of insoluble anticancer chemotherapeutics curcumin in forms of CNs (≈15nm) and iRGDCNs (≈18 nm) were successfully prepared using a brilliant dropletcryodesiccation- driven crystallization approach. In addition to remarkable high drug loading capacity, superficially coated Pluronic F127 and iRGD conjugation endowed these curcumin nanocrystallites with excellent monodispersibility, stealth effect in systemic circulation, as well as tumor targeting ability. Taking advantage of these superior features, Pluronic F127 and iRGD contribute to the prolonging circulation time, improving tumor accumulation, and facilitating tumor uptake, which were superior to the exhibited by the traditional non-biological materials modification strategies, without eliciting any unfavorable adverse effects. Given that our nanocrystallite preparation strategy can be easily applied on other insoluble drugs, the current work will lead to a new avenue for development of anticancer chemotherapeutics.
References
- Yunpeng Ye, Lei Zhu, Ying Ma, Gang Niu, Xiaoyuan Chen. Synthesis and evaluation of new iRGD peptide analogs for tumor optical imaging. Bioorganic & medicinal chemistry letters. 2011; 21: 1146-1150.
- Dezhi Ni, Hui Ding, Shan Liu, Hua Yue, Yali Bao, Zhenhua Wang, Zhiguo Su, Wei Wei, Guanghui Ma. Superior Intratumoral Penetration of Paclitaxel Nanodots Strengthens Tumor Restriction and Metastasis Prevention. Small. 2015; 11: 2518-2526.
- Zhang Xiong, Li Yuebin, Cai Yaxuan, Zhang Yi, Tian Yiqun, Wang Lin, Gu Haoshuang, Chen Wei. Pluronic®F127 Micelles Template Biomimetic Assembly and Optical Properties of Red Fluorescent Silica Nanocapsules. Science of Advanced Materials. 201; 9: 608-615.
- Andre Lamont Thompson, Brian James Love. Thermodynamic properties of aqueous PEO–PPO– PEO micelles of varying hydrophilicity with added cisplatin determined by differential scanning calorimetry. Journal of Thermal Analysis and Calorimetry. 2017; 127: 1583-1592.
- Tarun Ojha, Vertikal Pathak, Yang Shi, Wim E. Hennink, Chrit T.W. Moonen, Gert Storm, Fabin Kiessling, Twan Lammers. Pharmacological and Physical Vessel Modulation Strategies to Improve EPR-mediated Drug Targeting to Tumors. Advanced Drug Delivery Reviews. 2017; 119: 44-60.
- Marta Overchuk, Gang Zheng. Overcoming obstacles in the tumor microenvironment: Recent advancements in nanoparticle delivery for cancer theranostics. Biomaterials. 2018; 156: 217-237.
- Hui Ding, Yanjuan Cai, Lizeng Gao, Minmin Liang, Beiping Miao, Hanwei Wu, et al. Exosome-like Nanozyme Vesicles for H2O2-responsive Catalytic Photoacoustic Imaging of Xenograft Nasopharyngeal Carcinoma. Nano letters. 2019; 1: 203-209.
- Xingli Cun, Jiantao Chen, Shaobo Ruan, Li Zhang, Jingyu Wan, Qin He, Huile Gao. A Novel Strategy through Combining iRGD Peptide with Tumor- Microenvironment-Responsive and Multistage Nanoparticles for Deep Tumor Penetration. ACS applied materials & interfaces. 2015; 7: 27458-27466.
- Dimakatso Alice Senthebane, Arielle Rowe, Nicholas Ekow Thomford, Hendrina Shipanga, Daniella Munro, Mohammad A M AL Mazeedi, Ham A, Kallmeyer K, Dandara C, Pepper MS. The Role of Tumor Microenvironment in Chemoresistance: To Survive, Keep Your Enemies Closer. International Journal of Molecular Sciences. 2017; 18: 1586.
- Yunlu Dai, Can Xu, Xiaolian Sun, Xiaonyuan Chen. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chemical Society reviews. 2017; 46: 3830-3852.
- Matthew R. Dreher, Wenge Liu, Charles R. Michelich, Mark W. Dewhirst, Fan Yuan, Ashutosh Chilkoti. Tumor vascular permeability, accumulation, and penetration of macromolecular drug carriers. Journal of the National Cancer Institute. 2006; 98: 335-344.
- Vikash P. Chauhan, Triantafyllos Stylianopoulos, John D. Martin, Zoran Popovic, Ou Chen, Wild S. Kamoun, it al. Normalization of tumour blood vessels improves the delivery of nanomedicines in a size-dependent manner. Nature nanotechnology. 2012; 7: 383-388.
- Miriam Colombo, Steven Orthmann, Marco Bellini, Sven Staufenbiel, Roland Bodmeier. Influence of Drug Brittleness, Nanomilling Time, and Freeze- Drying on the Crystallinity of Poorly Water-Soluble Drugs and Its Implications for Solubility Enhancement. Aaps Pharmscitech. 2017; 18: 2437-2445.
- Bo Sun, Yoon Yeo. Nanocrystals for the parenteral delivery of poorly watersoluble drugs. Current Opinion in Solid State & Materials Science. 2012; 16: 295-301.