
Research Article
Austin J Cancer Clin Res. 2022; 9(1): 1099.
CXCR4 is Involved in the Regulation of the Functions of c-Src in RCC Development
Yan R1#, Pang Q1#, Chen J2#, Shi J3*, Gan X1* and Wang L1*
1Department of Urology, Changhai Hospital, Naval Medical University (Second Military Medical University),Shanghai, People’s Republic of China
2Department of Urology, Henan Provincial Corps Hospital of Chinese People’s Armed Police, Zhengzhou, People’s Republic of China
3Department of Urology, Changzheng Hospital, Naval Medical University (Second Military Medical University), Shanghai, People’s Republic of China
#Co First Author
*Corresponding author: Jiazi Shi, Department of Urology, Changzheng Hospital, Naval Medical University (Second Military Medical University), Shanghai, 200003, People’s Republic of China
Xinxin Gan, Department of Urology, Changhai Hospital, Naval Medical University (Second Military Medical University), Shanghai, 200433, People’s Republic of China
Linhui Wang, Department of Urology, Changhai Hospital, Naval Medical University (Second Military Medical University), Shanghai, 200433, People’s Republic of China
Received: January 25, 2022; Accepted: February 21, 2022; Published: February 28, 2022
Abstract
Renal cell carcinoma is a common malignant urinary tumor and CXCR4 plays an important role in the development of renal cell carcinoma. However, the role of c-Src gene in the development of renal cell carcinoma is still unclear. In this study, we found that CXCR4 can directly bind to SRC and CXCR4 is involved in the regulation of c-Src expression. C-Src is highly expressed in renal cell carcinoma and promotes cell division, proliferation, invasion and reduces apoptosis in renal cell carcinoma. Highly expressed c-Src is associated with poor prognosis in patients with RCC, and affects the infiltration of CD4+ T cells and macrophages in the tumor microenvironment. In addition, high SRC expression is associated with the expression of multiple immune checkpoints, high tumor mutation burden and high microsatellite instability which indicates the potential of SRC to predict the response to the immune checkpoint block therapy.
Keywords: CXCR4; SRC; Renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is one of the most common urinary tumors, causing over one hundred thousand deaths around the world annually [1,2]. With the develop of imaging technology, more and more renal tumors were found out in an early stage. However, the lack of obvious symptoms and signs still leads to metastatic disease in many patients when they were diagnosed. Targeted drugs, such as sunitinib, sorafenib and pazopanib, have significantly improved the prognosis of patients with metastatic renal cell carcinoma (mRCC) since 2006 [3-5]. With the surprising occurrence of immune checkpoint inhibitor, combined therapy has become the very first choice for mRCC patients [6,7]. However, in face of the malignant tumors, the effect of drug treatment is limited, so further explore of the develop mechanism of tumor is necessary.
CXC chemokine receptor type 4 (CXCR4) is an important molecule that highly expressed in a variety of cancers [8-10]. It was initially found to act as a co-receptor of T-tropic HIV viruses to entry CD4+ T cells [11]. CXCR4 binds to its ligand CXC motif chemokine ligand 12 (CXCL12), also known as Stromal cell-derived factor-1 (SDF-1), and then activate multiple pathways, such as PI3K/AKT pathway and Ras/Raf pathway, which lead to the enhance of diverse tumor biological behaviors, including proliferation, migration, invasion and transcriptional activation [12-14]. In our previous study, we found that non-muscle myosin heavy chain-IIA (NMMHC-IIA) and hypoxia-inducible factor-1α (HIF-1α) can interact with CXCR4 [15,16]. However, potential interacting proteins and concrete mechanisms need to be explored to help a more comprehensive understanding of the function of CXCR4.
The proto-oncogene c-Src is a member of Src family kinases (SFKs), a nine-gene family of non-receptor tyrosine kinases, consisting of SRC, YES, FYN, LCK, HCK, FGR, LYN, BLK and YRK [17-21]. As a cytoplasmic protein tyrosine kinase, SRC interacts with many focal-adhesion proteins, adaptor proteins and transcription factors in PI3K, MAPK, STAT3, FAK signaling pathways which contributes to its important role in modulation of cytoskeletal organization, angiogenesis, invasion, cell cycle progression and proliferation [17,22]. However, its function in the development of renal cell carcinoma remains to be explored.
In this study, the relationship between CXCR4 and c-Src and the role of c-Src in the development of renal cell carcinoma were explored.
Materials and Methods
Clinical RCC tissue samples
The study design was approved by the Changzheng Hospital Ethics Committee, and informed consent was obtained from each patient. We obtained 29 pairs of tumor and adjacent tissue samples and 98 tumor tissue samples from patients with RCC underwent nephrectomy, partial or radical, at Changzheng Hospital, Naval Medical University, Shanghai, China.
Cell lines and culture conditions
Human RCC cell line (A498 and ACHN) was obtained from the Chinese Academy of Sciences (Shanghai, China) and underwent STR authentication. The cells were incubated in DMEM high glucose medium (Gibco) containing 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (Gibco). All cells were grown as a monolayer on the plastic cell culture dishes at 37°C in a humidified atmosphere containing 5% CO2.
Lentiviral vectors and infection
The lentivirus encoding CXCR4, sh-CXCR4, sh-c-Src plasmids were packaged at GenePharma (Shanghai, China) and cells were infected following the manufacturer’s instruction.
RNA isolation and RT-PCR
Total RNA was extracted from fresh-frozen tissues or cultured cells using RNAiso Plus (Takara) and then underwent reverse transcription (Takara, RR036A) on Veriti Dx platform (Applied Biosystems) and RT-PCR (Takara, RR420A) on StepOne platform (Applied Biosystems). Relative mRNA was analyzed using 2-ΔΔCT method.
Western blotting
Total protein from fresh-frozen tissues or cultured cells was harvested with ice-cold Pierce IP (Thermofisher) and protease inhibitor cocktail (Beyotime Biotechnology). Equivalent amounts of protein were separated on 10% SDS-PAGE (Epizyme Biotech), transblotted onto 0.22μm Amersham Protran NC membrane (Cytiva). Membranes were blocked with 5% skimmed milk in TBST buffer for 2 hours at 4oC and incubated with the indicated primary antibodies with gentle rocking overnight at 4°C. Membranes were further incubated with the corresponding HRP-conjugated secondary antibodies (Beyotime Biotechnology) for 2 hours at 4°C. Finally, protein bands were imaged using enhanced chemiluminescence detection kit (Epizyme Biotech) and analyzed with the chemiluminescence imager Chemi Doc XRS+ (BIO-RAD).
Coimmunoprecipitation
Co-IP was performed as the manufacturer’s instructions (Pierce Co-Immunoprecipitation (Co-IP) Kit, Thermfisher). Protein complex pull-down with CXCR4 antibody from total protein harvested from ACHN cells. The CXCR4-pull-down products were subjected to 10% denaturing polyacrylamide gel electrophoresis and visualized by silver staining. The protein bands were analyzed using MS/MS spectra and the results were re-confirmed by another co-immunoprecipitation using CXCR4 antibody and SRC antibody.
Immunohistochemistry
Paired tumor and adjacent paraffin tissue samples were cut into slices and conducted a common immunohistochemistry procedure with primary CXCR4 or SRC antibody (Proteintech) and appropriate secondary antibody.
Flow cytometry
The reagents used to detect cell cycle and apoptosis by flow cytometry were purchased from BD Pharmingen and cells were tested according to the manufacturer’s standard instructions.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded into 96-well culture plates (2x10³ cells per well). At indicated time, 10μl CCK-8 reagent (Dojindo Molecular Technologies) was added to each well and incubated for 2h at 37°C. Absorbance values at a wavelength of 450nm were recorded using a SpectraMax Paradigm microplate reader (Molecular Devices). Viability (%) was calculated based on the optical density (OD) values as follows: (OD of time sample - blank)/(OD of 0h control sample - blank) x 100%.
Transwell invasion assay
The transwell invasion assay were performed with the 8μm cell culture insert (BD Falcon) and Matrigel matrix (Corning) following the manufacturer’s instructions. Cells were stained with 0.5% crystal violet.
Bioinformatics analysis
SRC expression in RCC was examined using the TIMER and UALCAN databases. The LinkedOmics database was used to study the signaling pathways related to SRC expression. Networks were generated using the STRING. TIMER and xCell were used to analyze the correlations among tumor-infiltrating immune cells. All analysis methods and R package were implemented by R foundation for statistical computing (2020) version 4.0.3 and software packages ggplot2 and pheatmap.
Statistical analysis
Statistical analysis was performed using the GraphPad Prism version 8.0 (GraphPad software, USA) and Statistical Package for Social Sciences (SPSS 17.0 for Windows, SPSS, Chicago, IL). The measurement data are presented as mean ± SD. An independent samples t test was used to analyze the differential expression levels of c-Src mRNA between the RCC tissues and the adjacent normal tissues from TCGA databases. Correlations between c-Src expression and clinicopathological characteristics were analyzed by the Pearson’s Chi squared test. Overall survival (OS) analysis was performed by Kaplan-Meier plots and the differences were compared using the log-rank test. Univariate and multivariable analyses were performed using the Cox proportional hazards regression models. A two-tailed P value of 0.05 was considered statistically significant.
Results
SRC directly combine with CXCR4 and locates downstream CXCR4
In order to find potential proteins that interact with CXCR4, a coimmunoprecipitation (co-IP) was conducted using CXCR4 antibody and lysates of ACHN cells. The result showed the capacity of CXCR4 to bind to various proteins and the 60 kDa band was identified as c-Src (Figure 1A). A similar conclusion was reached by another two co-immunoprecipitation tests using SRC antibody to pull down protein complex in lysates of A498 cells and CXCR4 antibody to pull down protein complex in lysates of ACHN cells separately (Figure 1B). To clarify the relationship between CXCR4 and SRC, we detected the expression of CXCR4 and SRC in 29 pairs of tumor tissue and adjacent tissue using immunohistochemistry. The results showed that both SRC and CXCR4 were highly expressed in RCC tissue (Figure 1C). Statistical analysis showed that the expression of SRC was positively correlated with the expression of CXCR4 (Figure 1D). Next, we considered whether c-Src locates upstream or downstream CXCR4. In order to clarify their relationship, we constructed A498 cell lines knocking-down CXCR4 and c-Src respectively, and detected the mRNA expression levels of the two genes by RT-PCR. It showed that the knockdown of CXCR4 significantly reduced the expression of c-Src and knockdown of c-Src did not affect the expression of CXCR4 on the contrary (Figure 1E). Therefore, we drew a conclusion that c-Src locates downstream CXCR4.
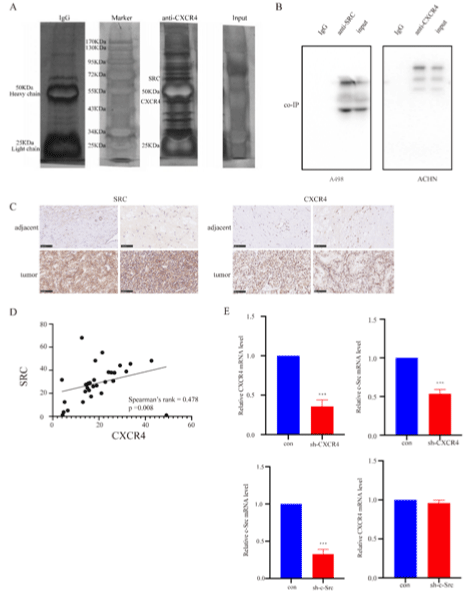
Figure 1: CXCR4 interacts with SRC and is involved in the regulation of c-Src.
A) The co-IP result using CXCR4 antibody in ACHN cells. B) The co-IP results using SRC antibody in A498 cells and CXCR4 antibody in ACHN cells. C) The results
of immunohistochemistry in paired RCC tumor and adjacent tissues. D) The correlation between the expression level of SRC and CXCR4 in immunohistochemistry.
E) The RT-PCR results after CXCR4 and c-Src knockdown in A498 cells.
C-Src highly expressed in RCC and indicates poor prognosis
To clarify the role of c-Src in the development of renal cell carcinoma, we first checked the expression of c-Src in RCC tissues by Western-Blotting and RT-PCR. The results showed that c-Src is highly expressed in RCC tissues (Figure 2A and 2B). Patients with high c-Src expression had a poorer overall survival rate according to our own follow-up data (Figure 2C). Bioinformatics analysis is used, and the results are just consistent with our conjectures. C-Src was highly expressed in the ccRCC cancer tissues compared with normal tissues (Figure 2D). Also, the expression of c-Src was higher in clinical late-stage and M1 stage tumors (Figure 2E and 2F), and high c-Src expression leads to a worse prognosis (Figure 3D).

Figure 2: c-Src is highly expressed in RCC and leads to a worse prognosis.
A) The protein level of SRC in tumor tissues and adjacent tissues of clinical RCC patients. B) The mRNA level of c-Src in tumor tissues and adjacent tissues of
clinical RCC patients. C) The OS of clinical RCC patients with different c-Src expression level. D) The mRNA level of c-Src in tumor tissues and normal tissues
of RCC patients from TCGA database. E) The mRNA level of c-Src in tumor tissues of RCC patients with different clinical T stages from TCGA database. F) The
mRNA level of c-Src in tumor tissues of RCC patients with different clinical M stages from TCGA database. G) The OS of clinical patients with different c-Src
expression level from TCGA database.
CXCR4 is involved in the regulation to the functions of c-Src in RCC development
In order to clarify the role of high c-Src expression in the development of RCC and the regulatory role of CXCR4 on c-Src, c-Src knockdown, CXCR4 overexpression and simultaneously CXCR4 overexpression and c-Src knockdown A498 cell lines were constructed by lentivirus infection. The cell line constructions were verified by RT-PCR and Western Blotting (Figure 3A and 3B). Then flow cytometry test, CCK-8 assay and transwell invasion assay were progressed to check the influences on functions of RCC cell. The results showed that knockdown of c-Src significantly reduces cell division, cell proliferation and cell invasion and increases cell apoptosis (Figure 3C-3F). By contrast, overexpression of CXCR4 can increase cell division, cell proliferation and cell invasion but reduce cell apoptosis (Figure 3C-3F). And overexpression of CXCR4 can rescue cells from the inhibition on cell division, cell proliferation and cell invasion and promotion on cell apoptosis causing by knockdown of c-Src (Figure 3C-3F).
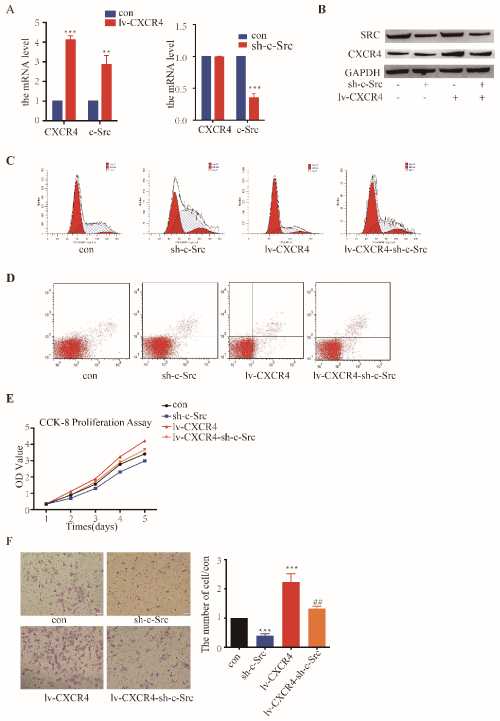
Figure 3: CXCR4 regulates the functions of c-Src in the development of RCC.
A) The mRNA expression after knockdown of c-Src and overexpression of CXCR4. B) The protein expression after knockdown of c-Src and overexpression of
CXCR4. C) The results of cell division cycle detected by flow cytometry. D) The results of cell apoptosis detected by flow cytometry. E) The results of CCK-8
proliferation assay. F) The results of transwell cell invasion assay.
SRC promote the development of RCC by affecting the tumor microenvironment
For a deeper sense of understanding on the function of c-Src in the development of RCC, we explored the interaction proteins of SRC by bioinformatics. The protein-protein interaction network (PPI network) displayed the top 45 proteins associated with SRC (Figure 4A). The GO and KEGG enrichment showed that those proteins join mainly in the ionic transportation and pH regulation (Figure 4B).
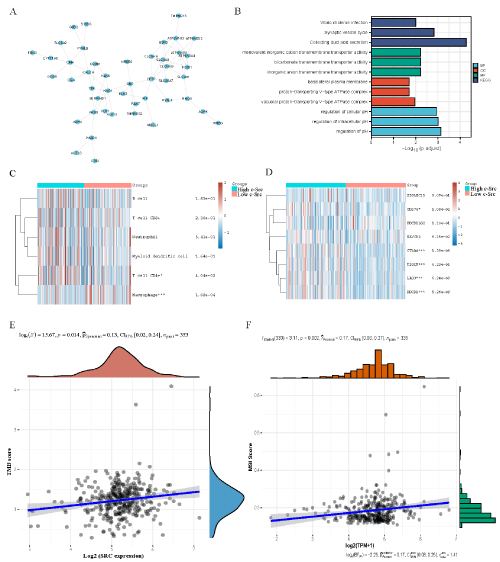
Figure 4: c-Src is associated with the changes of tumor microenvironment in RCC.
A) The PPI network of the top 45 proteins associated with SRC. B) The GO and KEGG enrichment of the 45 associated proteins. C) The immune infiltration level
in tumor tissues with different c-Src expression. D) The immune checkpoint level in tumor tissues with different c-Src expression. E) The correlation between tumor
mutation burden score and c-Src expression. F) The correlation between microsatellite instability score and c-Src expression.
With the emergence of immune checkpoint block (ICB) therapy, the evaluation of tumor immune infiltration status in RCC patients gets more essential and may improve the treatment response rate. From the results of bioinformatics analysis, we found that SRC was correlated with low infiltration levels of macrophages (p=1.68e-04) and high infiltrating levels of CD4+ T cells (p=4.04e-02) (Figure 4C). Also, SRC was significantly correlated with high expression of immune checkpoints, such as CD274 (p=1.08e-02), CTLA4 (p=1.39e-06), TIGIT (p=6.23e-05), LAG3 (p=6.24e-09), and PDCD1 (p=5.26e-09) (Figure 4D). When analyzing the correlation between SRC expression and tumor mutation burden (TMB) and microsatellite instability (MSI), we found that SRC expression was positively correlated with TMB (P=0.014) and MSI (P=0.002) (Figure 4E and 4F), suggesting that c-Src may predict the clinical response to ICB in RCC patients.
Discussion
Because of the hidden onset and insensitivity to radiotherapy and chemotherapy, advanced RCC remains a common fatal malignant tumor [23-25]. Although the emergence of targeted drugs improves the prognosis of patients with advanced renal cell carcinoma and the combination with immune checkpoint inhibitors further improves its efficacy, the overall benefit is still limited [3-7]. Therefore, it is still necessary to study the development mechanism of RCC.
In our previous research, functions of CXCR4 in RCC development were studied. We found that CXCR4 nuclear localization after binding to its ligand SDF-1 occurs in metastatic tissues but not the primary lesion [26]. Then, we found that CXCR4 knockdown inhibits cell growth and metastasis in RCC cells [27]. In order to learn more about the influence of the nuclear localization of CXCR4, we first identified the nuclear localization sequence of CXCR4 in RCC by constructing expression plasmids of different deletants [28]. Then we found nuclear localization of CXCR4 is dependent on NMMHCIIA in RCC [15]. Most importantly, we found that CXCR4 and hypoxia-inducible factor-1α (HIF-1α) colocalized in RCC cells and interacted with each other which formed a positive feed-forward loop to promote the development of RCC [16].
In this study, we found that CXCR4 can directly combine with SRC. This makes us wonder whether CXCR4 is involved in regulating the expression of c-Src. We found that CXCR4 knockdown significantly reduced the expression of c-Src but not vice versa. Therefore, we think c-Src may be a downstream molecule of CXCR4. The knockdown of c-Src significantly reduced cell division, cell proliferation and cell invasion and increased cell apoptosis. And overexpression of CXCR4 can rescue cells from the inhibition of cell division, cell proliferation and cell invasion and promotion of cell apoptosis causing by knockdown of c-Src. Thus, CXCR4 is involved in regulating the role of c-Src in promoting the development of RCC.
The results of bioinformatics analysis verified some of our conclusions and found that c-Src may play a role in regulating tumor microenvironment. We found that the expression level of c-Src affects the infiltration of immune cells in the tumor microenvironment and is related to the expression of multiple immune checkpoints which may indicate that c-Src can be a biomarker in screening patients suitable for immune checkpoint combined therapy.
Interaction between CXCR4 and c-Src was observed in many other cancers, such as breast cancer, prostate cancer and gastric cancer [29-32]. However, most of their studies only indirectly show that CXCR4 play a regulatory role to SRC. In our study, we found that SRC is one of the proteins directly bind to CXCR4 and is regulated by CXCR4 at both mRNA and protein levels in RCC. But the specific regulation mechanism of how CXCR4 influence the expression level of c-Src remains to be further studied to make clear how CXCR4 regulates the transcription of c-Src and what’s the effect of direct binding on their own protein functions.
The phenomenon that Src expression was associated with immune cell infiltration was found in both benign and malignant diseases [20,33-35]. But the mechanism behind this phenomenon still needs further experimental proofs. There’s a bioinformatics study showed that the differential expression of CXC kinase family affects the immune infiltration of RCC, and SRC family is the downstream target of CXC kinase family. This is consistent with our conclusion. However, the effects of SRC on the microsatellite instability score, tumor mutation burden score and the efficacy of immune checkpoint inhibitor therapy need to be further explored in clinical trials.
Overall, our study found that CXCR4 can directly bind to SRC and regulate the function of c-Src in RCC development. C-Src is involved in the regulation of immune infiltration in tumor microenvironment and affects tumor MSI and TMB score. It has the potential to be used as a screening marker of immune checkpoint inhibitors.
Data Availability Statement
All data, models, and code generated or used during the study appear in the submitted article.
References
- Padala SA, et al. Epidemiology of Renal Cell Carcinoma. World journal of oncology. 2020; 11: 79-87.
- Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68: 394-424.
- Motzer RJ, et al. Sunitinib in patients with metastatic renal cell carcinoma. JAMA. 2006; 295: 2516-2524.
- Escudier B, et al. Sorafenib for treatment of renal cell carcinoma: Final efficacy and safety results of the phase III treatment approaches in renal cancer global evaluation trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009; 27: 3312-3318.
- Sternberg CN, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010; 28: 1061- 1068.
- Rini BI, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med. 2019; 380: 1116-1127.
- Motzer RJ, et al. Avelumab plus Axitinib versus Sunitinib for Advanced Renal- Cell Carcinoma. N Engl J Med. 2019; 380: 1103-1115.
- Chen IX, et al. Blocking CXCR4 alleviates desmoplasia, increases T-lymphocyte infiltration, and improves immunotherapy in metastatic breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 2019; 116: 4558-4566.
- Wang D, et al. Exosome-encapsulated miRNAs contribute to CXCL12/ CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer letters. 2020; 474: 36-52.
- Jäger B, et al. CXCR4/MIF axis amplifies tumor growth and epithelialmesenchymal interaction in non-small cell lung cancer. Cellular signaling. 2020; 73: 109672.
- Feng Y, Broder CC, Kennedy PE & Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science (New York, N.Y). 1996; 272: 872-877.
- Teicher BA & Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010; 16: 2927-2931.
- Janssens R, Struyf S & Proost P. The unique structural and functional features of CXCL12. Cellular & molecular immunology. 2018; 15: 299-311.
- Daniel SK, Seo YD & Pillarisetty VG. The CXCL12-CXCR4/CXCR7 axis as a mechanism of immune resistance in gastrointestinal malignancies. Seminars in cancer biology. 2020; 65: 176-188.
- Xu Z, et al. NMMHC-IIA-dependent nuclear location of CXCR4 promotes migration and invasion in renal cell carcinoma. Oncology reports. 2016; 36: 2681-2688.
- Bao Y, et al. A feed-forward loop between nuclear translocation of CXCR4 and HIF-1α promotes renal cell carcinoma metastasis. Oncogene. 2019; 38: 881-895.
- Patel A, Sabbineni, H, Clarke A & Somanath PR. Novel roles of Src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life sciences. 2016; 157: 52-61.
- Anguita E & Villalobo A. Src-family tyrosine kinases and the Ca2+ signal. Biochimica et biophysica acta. Molecular cell research. 2017; 1864: 915-932.
- Jin W. Regulation of Src Family Kinases during Colorectal Cancer Development and Its Clinical Implications. Cancers. 2020; 12.
- Caner A, Asik E & Ozpolat B. SRC Signaling in Cancer and Tumor Microenvironment. Advances in experimental medicine and biology. 2021; 1270: 57-71.
- Matozaki T, Kotani T, Murata Y & Saito Y. Roles of Src family kinase, Ras, and mTOR signaling in intestinal epithelial homeostasis and tumorigenesis. Cancer science. 2021; 112: 16-21.
- Belli S, et al. c-Src and EGFR Inhibition in Molecular Cancer Therapy: What Else Can We Improve? Cancers. 2020; 12.
- Hsieh JJ, et al. Renal cell carcinoma. Nature reviews. Disease primers. 2017; 3: 17009.
- Motzer RJ, et al. NCCN Guidelines Insights: Kidney Cancer, Version 1.2021. Journal of the National Comprehensive Cancer Network: JNCCN. 2020; 18: 1160-1170.
- Ljungberg B, et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2019 Update. Eur Urol. 2019; 75: 799-810.
- Wang L, et al. CXCR4 nuclear localization follows binding of its ligand SDF- 1 and occurs in metastatic but not primary renal cell carcinoma. Oncology reports. 2009; 22: 1333-1339.
- Wang L, et al. Silencing of CXCR4 by RNA interference inhibits cell growth and metastasis in human renal cancer cells. Oncology reports. 2012; 28: 2043-2048.
- Wang LH, et al. Identification of nuclear localization sequence of CXCR4 in renal cell carcinoma by constructing expression plasmids of different deletants. Plasmid. 2010; 63: 68-72.
- De Luca A, et al. Src and CXCR4 are involved in the invasiveness of breast cancer cells with acquired resistance to lapatinib. Cell cycle (Georgetown, Tex.). 2014; 13: 148-156.
- Conley-LaComb MK, et al. Pharmacological targeting of CXCL12/CXCR4 signaling in prostate cancer bone metastasis. Mol Cancer. 2016; 15: 68.
- Cheng Y, et al. CXCL12/SDF-1α induces migration via SRC-mediated CXCR4-EGFR cross-talk in gastric cancer cells. Oncology letters. 2017; 14: 2103-2110.
- Zhao M, Discipio RG, Wimmer AG & Schraufstatter IU. Regulation of CXCR4- mediated nuclear translocation of extracellular signal-related kinases 1 and 2. Molecular pharmacology. 2006; 69: 66-75.
- Xiu MX, Liu YM, Chen GY, Hu C & Kuang BH. Identifying Hub Genes, Key Pathways and Immune Cell Infiltration Characteristics in Pediatric and Adult Ulcerative Colitis by Integrated Bioinformatic Analysis. Digestive diseases and sciences. 2021; 66: 3002-3014.
- Liu ST, Pham H, Pandol SJ & Ptasznik A. Src as the link between inflammation and cancer. Frontiers in physiology. 2013; 4: 416.
- Xu W, et al. Multi-omics reveals novel prognostic implication of SRC protein expression in bladder cancer and its correlation with immunotherapy response. Annals of medicine. 2021; 53: 596-610.