
Editorial
Austin Chromatogr. 2015;2(1): 1025.
Lipid Molecular Profiling of Nowadays and Future
Chuan-Ho Tang1,2*
1National Museum of Marine Biology and Aquarium, Taiwan
2Institute of Marine Biodiversity and Evolutionary Biology, National Dong Hwa University, Taiwan
*Corresponding author: Chuan-Ho Tang, National Museum of Marine Biology and Aquarium and Institute of Marine Biodiversity and Evolutionary Biology, National Dong Hwa University, Taiwan
Received: January 12, 2015; Accepted: January 20, 2015; Published: January 20, 2015
Editorial
Organisms dynamically apply the intrinsic polymorphism of lipids and the involved molecular interactions, such as lipid-lipid and lipid-protein, to create a variety of biological complexes, such as cellular membranes, lipid domains, and lipoproteins, with the unique compositions maintaining multiple functionalities for life requirements. Progress in the analytical technologies, biophysics, chemical biology etc. has attracted attention to the mechanistic description of such complexes functioning in molecular and physical terms. Effects of lipid molecular structure on assembly and physical properties of bio-complexes, such as cellular membrane of structure, tension, fluidity, permeability etc., and action and activity of protein, can therefore be related; thus cell of operation, regulation, and dysfunction should be accompanied a suited or altered lipid composition [1-3]. For understanding or diagnosing an integral whole of cellular function, it is essential to have insight into the lipids composition in molecular structure level. Glycerophospholipids (GPs) are for example the major structural lipid in eukaryotic organisms. They are composed of a glycerol backbone with a phosphoryl-head group at the sn-3 position and a fatty acid substituent at the sn-1 and/ or sn-2 position as shown in Figure 1. Adopted GPs classification divides them into several classes, such as glycerophosphocholine, glycerophosphoethanolamine, glycerophosphoserine, glycerophosphoglycerol, and glycerophosphoinositol, according as the difference of phosphoryl-head group. Based on the substituent of fatty acid as ester, alkyl ether or vinyl ether linkage at the sn-1 position, each GP class can be further divided into the subclasses of phosphatidylcholine, plasmanylcholine, and plasmenylcholine. Another type of GP is the lyso-GP, which contain one glycerol hydroxyl substituent at either sn-1 or sn-2 position rather than a fatty acid substituent. The chain length and the double bond position of fatty acid substituent also contribute to the diversity of the GP molecular species. The complexities in the molecular species of lipids can be attributed to the diverse functions that biological system requires the lipids to perform. Such a kaleidoscopic possibility has supported an ongoing interest in life-related science, and thus offers a big challenge in the lipid analysis.
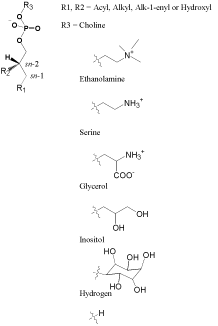
Figure 1: Chemical structures of glyserophospholipids.
Currently, the research methods of lipidomics, mainly mass spectrometry-based, can accomplish high throughput lipid analysis with comprehensive characterization of molecular structure, especially for GPs, in biological system. Electrospray ion source is most often adopted ionization technique for mass spectrometrybased lipids analysis. All GPs are readily ionizable in the ionization process which could be analyzed as ionized form, and/or cationic/ anionic adducts of them. Formation and abundance of the molecular ion depends on the ionization mode, lipid class, and the presence of appropriate small ions. Analysis of specific GP requires insight into the patterns of collision-induced decomposition for their molecular ions generated in the ionization process. Varied patterns of the molecular fragmentation derived from each molecular ion can inform about specific clues for conducting integral profiling of lipid composition. An approach named “shotgun” lipidomics has been developed to globally analyze the molecular composition of lipids directly from the diluted crude extracts of biological samples without chromatographic separation. Shotgun lipidomics is a multistep method that uses an intra-source separation, multidimensional mass spectrometry, and array analysis [4]. Employing the properties of electro spray ion source, the molecular ion of specific GP is predominantly generated (separated out) by the modification of inflow character and switch of ionization mode. Mass spectrometric analyses are conducted in specific precursor ion or neutral loss scan according to the GP molecular structural features generated in the fragmentation processes to create a multidimensional array of mass spectra. Finally, the molecular species profiles of each GP class can be comprehensively identified and quantified through the array analysis based on a mass spectrum derived from lipid classification-specified fragment ion. Alternatively, diagnosing the lipid class-specified mass spectrum sifts interested signals to acquire the full spectra of fragmentation further for determining the lipids molecular structure can also be adopted. Although it is facile in data acquisition of shotgun lipidomics, the complexities in the mass spectra resulting from the overlapping of isotope and isobaric signals require iterative processing of deconvolution to produce a detailed molecular species profile. Because many ion signals are composed of isobaric lipid molecular species in the mass spectrum, further mass spectrometric experiments should be conducted to estimate their ratio. In addition, a restriction must be consider in ionization process that ion suppression and lipid-lipid interactions is probably induced by the excess of lipid content in the infusion sample.
Chromatographic separation combined mass spectrometric analyses have becoming a definite tendency for lipid profiling analysis. This combination makes the raw dataset a further resolution, and greatly reduces the possibility of interferences in the ionization process. Currently, both reversed- and normal- phase chromatography has been adopted for the separation in lipids profiling depending on the analysis strategy [5,6]. Based on hydrophilic interaction with packing material, such as silica and diol modified-silica, the mixed lipids can be sequentially eluted from normal-phase column according to their class and subclass. In addition to dependence on head group, fully separated lyso-fractions and some sub-fractionation as result of difference in fatty acid linkage are for example obtained in the GP chromatography. Normal-phase chromatography thus offers an opportunity for comprehensively surveying each class of lipid molecular composition in one single run of analysis. However, each presented peak in the chromatogram consists of multitudinous lipid molecular species requiring iterative deconvolution processing as mentioned in shotgun lipidomics. Reversed-phase chromatography possesses the most power for resolving the molecular species through the hydrophobic interaction with fatty acid chains of lipids. By which, the isobaric molecule of lipid species, even just a difference in the position of double bond within fatty acid chain, can also be separated, but it is lipid class-independent absolutely. Beyond the past, significantly, superior separation efficiency with increased sensitivity has been achieved as a result of recent advances in the chromatographic system. Namely so-called “ultra (high)” performance liquid chromatography, using a column packed with the “core-shell” or sub-2 μm diameter particles respectively operated at less than 6,000 or capable of more than 10,000 psi of pressure system [7,8]. Owing to high complexity presented in the total lipid extracts of biological system, such chromatographic systems surely be worthy of consideration to provide an excellent profile of lipid molecular species.
Integrating both characters of normal- and reversed- phase chromatography have been attempting to presents a two-dimensional separation for the comprehensive lipid profiling of the complex crude extracts [9,10]. This integration can makes an excellent orthogonality to offers a great opportunity to resolve lipid composition further in lipidomics analysis. However, those arrangements of chromatography paid separation efficiency and/or operating facility as the cost of additional resolution. In the near future, therefore, the combination of lipid class fractionation directly followed by the molecular species resolution of each lipid class probably become an arm in lipidomics study if the chromatographic efficiency and performing automaticity are continually improving.
References
- Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003; 44: 1071-1079.
- Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta. 2003; 1612: 1-40.
- Van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008; 9: 112-124.
- Vigh L, Escriba PV, Sonnleitner A, Sonnleitner M, Piotto S, Maresca B, et al. The significance of lipid composition for membrane activity: new concepts and ways of assessing function. Prog Lipid Res. 2005; 44: 303-344.
- Okazaki Y, Kamide Y, Hirai MY, Saito K. Plant lipidomics based on hydrophilic interaction chromatography coupled to ion trap time-of-flight mass spectrometry. Metabolomics. 2013; 9: 121-131.
- Retra K, Bleijerveld OB, van Gestel RA, Tielens AG, van Hellemond JJ, Brouwers JF. A simple and universal method for the separation and identification of phospholipid molecular species. Rapid Commun Mass Spectrom. 2008; 22: 1853-1862.
- Tang CH, Tsao PN, Chen CY, Shiao MS, Wang WH, Lin CY. Glycerophosphocholine molecular species profiling in the biological tissue using UPLC/MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2011; 879: 2095-2106.
- Tang CH, Tsao PN, Lin CY, Fang LS, Lee SH, Wang WH. Phosphorylcholine-containing lipid molecular species profiling in biological tissue using a fast HPLC/QqQ-MS method. Anal Bioanal Chem. 2012; 404: 2949-2961.
- Nie H, Liu R, Yang Y, Bai Y, Guan Y, Qian D, et al. Lipid profiling of rat peritoneal surface layers by online normal- and reversed-phase 2D LC QToF-MS. J Lipid Res. 2010; 51: 2833-2844.
- Lisa M, Cifkova E, Holcapek M. Lipidomic profiling of biological tissues using off-line two-dimensional high-performance liquid chromatography-mass spectrometry. J Chromatogr A. 2011; 1218: 5146-5156.