
Research Article
Austin J Clin Case Rep. 2021; 8(8): 1224.
Pilot Assessment of a Rapid Test for the Detection of COVID-19 Disease by Using Latex Agglutination
Cariaga-Martínez A1*, Gutiérrez KJ1 and Alelú-Paz R1,2*
1Laboratory of Neuroscience Elena Pessino Gómez del Campo, Madrid Scientific Park, Spain
2Department of Psychology, Universidad Francisco de Vitoria, Madrid, Spain
*Corresponding author: Alelú-Paz R, Department of Psychology, Universidad Francisco de Vitoria. Ctra. Pozuelo-Majadahonda KM 1.800. ZIP: 28223. Pozuelo de Alarcón, Madrid, Spain
Cariaga-Martínez A, Laboratory of Neuroscience Elena Pessino Gómez del Campo, Madrid Scientific Park, ZIP: 28049. Madrid, Spain
Received: June 08, 2021; Accepted: June 30, 2021; Published: July 07, 2021
Abstract
The rapid spread of the SARS-CoV-2 virus, which was declared a pandemic by the WHO in March 2020, has forced the scientific community to develop rapid detection tests in order to detect positive cases and implement the containment measures established in each country. In this regard, the techniques used (RTPCR, antibody test, etc.) have a number of drawbacks: require specialized personnel, in addition to, in some cases, obtaining results after 24 hours. Agglutination tests, widely used in the detection of viral particles, represent a simple, inexpensive and scalable method that would allow screening studies to be carried out in large populations. In this paper, we present a SARS-CoV-2 detection test based on this methodology, which could be considered as a complementary method to the techniques used for the detection of SARSCoV-2.
Keywords: COVID-19; SARS-CoV-2; Diagnosis; Rapid test; Agglutination
Introduction
The pandemic declared by WHO in March 2020 has resulted in thousands of deaths and a large expenditure of money in the development of rapid detection tests [1]. Initially, serological tests were used to assess the presence of infection although real-time PCR quickly established itself as a gold standard method [2]. Antigen tests were developed well into the pandemic and were blamed for lack of sensitivity and specificity. Nowadays, antigen tests have become popular, but their processing can only be carried out by competent healthcare personnel. In this sense, the dynamics of the pandemic has forced us to develop self-administered tests that allow us to obtain results in a very short time, in order to detect positive cases and implement the control measures established in each of the countries. As we have previously evaluated, one of the main difficulties in detecting the presence of SARS-CoV-2 infection is the technical difficulty in applying them (high costs and the need for highly qualified technical personnel), which makes it impossible to administer this type of test on a massive scale for screening studies. In this respect, the agglutination test is a simple, one-step method used for the detection of viral antigens in clinical specimens [3,4]. These assays are based on the initial fixation of specific antiviral antibodies on erythrocytes or latex particles which allows incubation with the clinical sample in which the antigen is being investigated and the particles agglutinate if the appropriate antigen is present. Although these tests require to use other techniques in order to confirm the results, due to the high percentage of nonspecific reactions, they can be used for screening studies in very large populations [5].
In this pilot test, we evaluated the technical and performance characteristics of a rapid SARS-CoV-2 detection test by using this methodology, that is, latex beads agglutination for application as an inexpensive, scalable, and complementary method of COVID-19 detection.
Materials and Methods
Subjects
Sixty samples were obtained from persons that presented to the Emergency Department of the Hospital Puerta de Hierro- Majadahonda in Madrid (Spain), with suspected SARS-CoV-2 infection, with presence or absence of symptoms. The patients were randomly selected, informed of the existence of a pilot trial and were asked to participate. All the enrolled patients received the appropriate information and signed the informed consent in accordance with the regulations for studies with human samples. The present study was positively evaluated and approved by the Hospital’s Bioethics Committee.
Samples
Saliva samples were collected after signing the informed consent and consisted of spontaneously generated saliva (at least 0.2ml) that was collected in sterile tubes and kept at -80°C until final processing. The Emergency Department and the Biobank Unit from the Hospital Universitario Puerta de Hierro Majadahonda assisted in the collection and preservation of the samples.
Real-time PCR
Patients’ samples were assessed by using the following Real-time PCR kits:
• Lyophilized 1-step RT-PCR Polymerase Mix (TIB MOLBIOL. Cat.- No. 90-9999-96).
• As an extraction control PCR (to verify the presence of amplifiable nucleic acids): LightMix Modular EAV RNA Extraction Control (Roche. Cat.- No. 66-0909-96).
• To detect SARS and SARS-CoV-2: LightMix Modular SARS-CoV (COVID19) E-gene (Roche. Cat. No.- 53-0776-96).
Briefly, the FastCycle Protocol: (Table 1).
Program Step
RT Step
Denaturation
Cycling
Cooling
Parameter
Analysis Mode
None
None
Quantification mode
None
Cycles
1
1
42-45
1
Target (°C)
50-55
95
95/60
40
Hold (hh:mm:ss)
0:03:00
0:00:30
00:00:03/00:00:12
0:00:10
Ramp Rate (°C/s) 96
4.4
4.4
4.4/2.2
1.5
Ramp Rate (°C/s) 384
4.6
4.6
4.6/2.4
2
Acquisition Mode
None
None
None/Single
None
Ct values higher than 40 were considered negative.
Table 1:
Antibody preparation
Fifty micrograms of recombinant (C-terminus) His-tagged ACE2 protein (Sino Biological. Ref: 10108-H08B) was mixed with 25ug of anti-His antibody (ThermoFisher. Ref: MA121315) and rotated ON at 4°C. Sterile 0.025M MES buffer (VWR. Cat.: E169) pH 6 was added to facilitate rotation (ON/4°C).
Commercially available 0.3-micron diameter latex beads (Sigma. Cat.: LB3-1ml) at a final concentration of 0.5% solids was used as support. Briefly, 20ul of commercial solution (10%) was taken and diluted to a final volume of 600ul with MES buffer +0.1% BSA (Sigma. Cat: A0336) +0.01% Triton X100 to block nonspecific interactions and as a surfactant to prevent self-aggregation of the beads.
The antigen-antibody (ACE2/antiHis) solution was placed in contact with the diluted support and rotated for 2 hours at RT. After incubation, aliquots of 100ul were centrifuged for 10 minutes at 6000xg to pellet the beads. Three washes with 500ul of 0.025M MES buffer +0.1% BSA were carried out. Finally, beads were resuspended with MES buffer to a final solid concentration of 0.5% and kept at 4°C. The complete protocol is detailed in supplementary materials.
Variable fine-tuning (prior to pilot test with human samples)
Given the size of the virus (approximately 0.1 microns), a “viral mock” was prepared by coating 0.1-micron latex beads (Sigma. Cat: LB1-1ml) with increasing amounts of recombinant viral spike protein (Sino Biological. Ref: 40589-V08B1). Selected variables (as antibody titration, reaction times, etc.) were tested against this viral mock and adjusted to obtain positive agglutination results in the range of 124 to 7.75 pg (considering that PCR detection is in the range of around 15pg). After obtaining this fine-tuning, the final protocol was defined, and the human samples kept at -80°C were used.
Performance of the assay
On a microscopy slide, we added 5ul of ACE-His sensitized beads solution in one spot and 5ul of 1% BSA sensitized beads (as a negative control) in another spot. To each group of beads, 5ul of saliva sample was added. The reactants were actively shaken in a circular motion with a sterile tip for 30 seconds and the reaction was allowed to proceed for up to 6 minutes.
Assay interpretation
In this assay, instantaneous agglutination in the negative control was related to the presence of strong interference in the sample. Therefore, if automatic agglutination was observed in the control spot, these samples were classified as indeterminate. On the other hand, actively homogenizing facilitates competition between the virions and other interferents in the sample. If the proportion of interferents is higher, disorganized agglutination will be observed when the saliva-beads interaction is forced. If the virions are the intermediary agents in the generation of the agglutination network, then the reaction will proceed more slowly and after 6 minutes a homogeneous agglutination pattern will be observed throughout the system (typically in a “starry sky” [6,7]). When compared to control, thus, these samples were classified as “positive”.
Statistical analysis
All the statistical analysis were carried out by using GraphPad Prism 8 software and a=0.05 was chosen as significance level.
Results and Discussion
In this paper, we present a pilot assessment of an antigen test based on particle agglutination, which generates interesting results with ample room for improvements. This test is inexpensive, easily scalable and the results’ interpretation require no more than six minutes and direct observation. Agglutination techniques are widely used, and, in this assay, we make use of the ability of the ACE2 protein to interact with the viral spicule. The presence of a commonly used tag (His tag) at the C-terminal end allows the ACE protein to bind to a specific anti-His antibody, leaving its site of interaction with the viral spicule free (N-terminal). Latex beads (commonly used in agglutination assays) preferentially capture the heavy chains of the antibodies, thus leaving exposed the protein portion of the ACE that will allow its interaction with the viral spicule that may be present in the sample. Also, placing an antibody as intermediary, moves the ACE molecule away from the latex bead thus avoiding steric hindrance and facilitating the exposure of the binding site. Consequently, if there are enough complete viral particles in the sample, a three-dimensional network may form and precipitate.
As a previous step, the selection of optimal conditions was carried out by using a “viral mock”: 0.1-micron latex beads sensitized with different amounts of the SARS-CoV-2 spike protein. ACE2-sensitized beads were put in contact and agglutination was observed. We also used this mock every time we were unable to use a positive sample in fine tuning the reaction overall. Table 2 indicates the semiquantitative results obtained with this procedure when spike protein quantity was assayed as a variable to mimic the “viral load”.
Total quantity of spike protein (ng)
124
62
31
15.5
7.75
Spike-Sensitized beads (μl)
10
10
10
10
10
ACE2-Sensitized beads (μl)
5
5
5
5
5
Results
+++
+++
+++
++
+
Table 2: Determination of optimal conditions for agglutination by using a “viral mock”.
Saliva, by its nature, is a type of sample that presents many interferents, thus in this assay, we increase the kinetics (by vigorous shaking) of the reaction to favor agglutination. If the interferents in the sample are in high proportion, rapid agglutination will be promoted, and these samples will be classified as indeterminate. If the interferents are not found in large proportion, the increase in kinetics will favor competition between interferents and viral particles, with specific interactions being slower to form but, due to the geometry of the virus itself, better established in space, so that the pattern will be specific (in a “starry sky” [6,7]) generating more stable networks although with longer formation time. These variables allow us to distinguish positive from negative samples, while the negative control avoids classifying samples derived from autoagglutination. Figure 1 shows a typical pattern of agglutination that is easily observed in a positive condition when compared to control spots. It also shows an indetermined reaction in comparison to control beads.
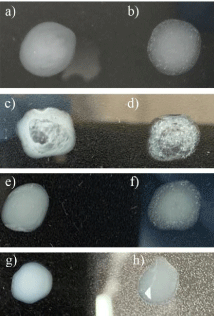
Figure 1: Typical results obtained from the rapid test for COVID detection.
a and c) Negative controls are beads sensitized with 1% BSA; b) Positive
reaction (homogeneous pattern of agglutination); d) Indeterminate reaction:
an immediate agglutination reaction is observed either in both the control and
the tested sample with a filamentous agglutination pattern. Patients’ samples.
e and g) Negative controls are beads sensitized with 1% BSA; f) Positive
reaction (homogeneous pattern of agglutination); h) Negative reaction (notice
the contrast to facilitate and ensure the observation of negative reactions in
patients’ samples).
After fine-tuning, ACE2-sensitized beads were assayed in human saliva samples. Of a total of 60 samples evaluated, 6 were indeterminate. The contingency Table 2 shows the obtained data, compared with real-time PCR data.
The data in the contingency table were evaluated with Fisher’s exact test, and statistically significant differences were observed (p-value=0.0441). Obtained sensitivity was 54.55% (95% CI=34.66 to 73 %) and obtained specificity was 75% (95% CI = 57.89 to 86.7 %) (Table 3).
Positive PCR
Negative PCR
Sensitivity
Specificity
Positive predictive value
Negative predictive value
Positive test
12
8
54.55%
75%
60%
70.59%
Negative test
10
24
95% CI = 34.66% to 73%
95% CI = 57.89% to 86.7%
95% CI = 38.66 to 78.1%
95% CI = 53.83 to 83.1%
Fisher’s Exacts test
p value<0.05 (p=0.0441)
Table 3: Contingency table for the pilot study.
The positive predictive value was 60% (95% CI = 38.66 to 78.1 %) and the negative predictive value was 70.59% (95%CI = 53.83 to 83.1 %). The overall power of the pilot test was 66.7%.
In this pilot test of 60 samples, we have achieved a relatively low sensitivity that can be explained by several causes. The main one, undoubtedly, is that the sensitivity of PCR to detect positive samples extends several weeks in time, thus the comparison of our test with a technique such as real time PCR is highly affected. In general, antigen tests depend on the presence of an active and complete virus, so detection peak is observed around one week after infection. For the type of test presented in this work, the comparison should probably be done within a specific time frame but given the pandemic conditions it has been very difficult to obtain highly controlled samples in terms of time from onset of symptoms or even the presence of symptoms.
The specificity data are more encouraging as they indicate that the test does indeed specifically find the virions present in a particularly complicated sample such as saliva. The high specificity also allows us to affirm that the number of false negatives would be low, thus fulfilling the criterion of a rapid screening test, and that more sensitive evaluations can be requested if necessary. On the other hand, for the diagnosis of a disease such as COVID-19, the presence of false positives always allows us to refer to confirmatory tests. A test like the present in this work, characterized by being very cheap and easily scalable, could be validated in a second stage to be used in mass events, schools, airports or any meeting requiring virtually instantaneous detection. Moreover, its low cost and ease of interpretation would allow self-administration and repetition over several points of times in case of indeterminacy of the results. We believe that it could be very useful as a complementary measure to the already established tests which are still prohibitively expensive for a large part of the population.
Acknowledgment
The authors would like to thank all those who have provided the samples (Dr. Antonio J. Sánchez, Dra. Rosa Capilla and Dra. Paqui Portero from the Hospital Universitario Puerta de Hierro Majadahonda and the Nava Handball Team, especially Dr. Carlos Navarro) and those who have worked to obtain them despite the difficulties encountered, with special mention to Álvaro Porras and Marcos Alberto Sanchez.
References
- Jee Y. WHO International Health Regulations Emergency Committee for the COVID-19 outbreak. Epidemiol Health. 2020; 42: e2020013.
- Younes N, Al-Sadeq DW, Al-Jighefee H, Younes S, Al-Jamal O, Daas HI, et al. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARSTotal CoV-2. Viruses. 2020;12: 582.
- Imwidthaya P, Egtasaeng C. Latex agglutination test for diagnosing cryptococcosis. J Med Assoc Thai. 1991; 74: 454-458.
- Wijedoru L, Mallett S, Parry CM. Rapid diagnostic tests for typhoid and paratyphoid (enteric) fever. Cochrane Database Syst Rev. 2017; 5: CD008892.
- Bhaskar S, Banavaliker JN, Hanif M. Large-scale validation of a latex agglutination test for diagnosis of tuberculosis. FEMS Immunol Med Microbiol. 2003; 39: 235-239.
- Mahat M, Adbullah W, Hussin C. Conventional Rapid Latex Agglutination in Estimation of von Willebran Factor: Method Revisited and Potential Clinical Applications. J Immunol Res. 2014; 2014: 850810.
- Medina MB, Shelver WL, Fratamico PM, Fortis L, Tillman G, Narang N, Cray WC Jr, Esteban E, Debroy A. Latex agglutination assays for detection of non-O157 Shiga toxin-producing Escherichia coli serogroups O26, O45, O103, O111, O121, and O145. J Food Prot. 2012; 75: 819-826.