1Department of Internal Medicine, Morsani College of Medicine, USA
2James A. Haley Veterans’ Hospital, USA
*Corresponding author: Jia-Wang Wang, Division of Allergy and Immunology, Department of Internal Medicine, University of South Florida Morsani College of Medicine, USA
Received: September 30, 2014; Accepted: October 30, 2014; Published: November 03, 2014
Citation: Reiser M, Li K, Lockey RF and Jia-Wang Wang. Lipopolysaccharide Responsive Beige-Like Anchor Subcellular Localization Involving in Vesicle Trafficking Responsive to Lipopolysaccharide. Austin J Clin Immunol. 2014;1(4): 1020. ISSN : 2381-9138
Lipopolysaccharide (LPS) responsive Beige-like Anchor (LRBA) gene is an important novel immune regulator, mutations of which cause common variable immunodeficiency, autoimmunity and inflammation. The underlying subcellular mechanism is unknown. Studies on LRBA subcellular localization and involvement in the dynamic vesicle trafficking may help to decipher its function of causing immune disorders. The present immune fluorescence confocal microscopy results show that LRBA is co-localized with Golgi proteins (GM-130, P-230 and GS-28), early endosome proteins [Early Endosome Antigen 1(EEA1), CLATHRIN, RAB4 and ADAPTIN-β], PKA subunits (RIIα, RIIβ and RIIC), and the microtubule protein, tubulin. Time lapse videos of live cells show LRBA-positive vesicles respond to LPS, which also stimulates LRBA nucleic translocation. This study demonstrates that LRBA is associated with the Golgi complex, endosomes, plasma membrane, nucleus, pseudopodia and microtubules, and vesicles. It suggests that LRBA plays anessential role in vesicle trafficking and signal transduction essential for normal immune response, especially against LPS-containing bacteria.
Keywords: LRBA; Subcellular localization; Vesicle trafficking; Lipopolysaccharide; Confocal microscopy
Toll is an essential receptor for host defense against fungal and Gram-positive bacterial infections in Drosophila [1]. The mammal Toll-Like Receptor 4 (TLR4) recognizes Lipopolysaccharides (LPS) from Gram-negative bacteria, fibrinogen cleaved by proteases from fungi and Gram-positive bacteria, and viral proteins (F protein from respiratory syncytial virus, vesicular stomatis virus glycoprotein G, poxviral protein A46) [2,3]. Therefore, this receptor is important to defend against Gram-negative and some Gram-positive bacteria [4], fungi [5] and viruses [6]. LPS is a crucial structural component of Gram-negative bacteria and a potent immunostimulators. Excessive LPS stimulation can result in systemic inflammation and death [7]. LPS-Responsive Beige-Like Anchor (LRBA) was initially identified as an LPS-up regulated gene in B cells [8,9] and has structural similarity to Lysosomal Trafficking regulator (LYST) and potentially is an A-Kinase Anchoring Protein (AKAP) [8]. It is composed of multiple domains and potentially may serve as a scaffold to interact with multiple proteins involved in the endomembrane system/ vesicle trafficking. However, the function of none of these domains has been determined and the exact function of LRBA is unknown [10]. Understanding the functions of LRBA became of increasing importance in 2012, when three research groups identified that six homozygous, functionally null germline mutations of LRBA cause an immune disease of Common Variable Immunodeficiency (CVID)with autoimmunity and inflammation, manifested as hypogammaglobulinemia, antibody deficiency, defective B-cell differentiation, recurrent infections, particularly respiratory infections, and variable autoimmune disorders including idiopathic thrombocytopenic purpura, autoimmune hemolytic anemia, and inflammatory bowel disease [11-13]. The average age of symptom onset is three years, earlier than the mean age of 26.3 years for most CVID [11,13-15]. In addition to immunological disorders, LRBA-deficient patients have an array of other medical problems which include: retarded growth, failure to thrive, growth hormone deficiency, asthma, monoarthritis, seizure disorders, granulomatous infiltration, finger clubbing, hepatosplenomegaly, allergic dermatitis, and nephrotic syndrome [11, 13,15]. These results demonstrate that LRBA is a unique CVID gene and plays a fundamental role in the immune system.
Despite these progresses, the exact subcellular and molecular mechanism of LRBA’s causing immune disorders remains unknown [16]. It is generally accepted that the subcellular location is closely related to physiological function of a protein and thus crucial to understanding its role in biological processes [17]. LRBA is implied as a vesicle trafficking regulator [10]. It is now realized that vesicle trafficking is crucial in transducing signals between cellular locations, as simple diffusion has a limited role in intracellular transport of signaling complexes [18]. About one-third of human proteins are delivered to the secretory and endocytic pathways through vesicle trafficking [19]. These proteins, to name a few, include proinflammatory cytokines, TNFα, IL-1 and IL-6, and their cellular receptors. Moreover, the Golgi Complex (GC) is viewed as the “headquarters” for signal transduction and cell-fate decisions, in addition to its “post office role” of cargo sorting/processing [20]. The 2013 Nobel Prize for vesicle trafficking research highlights the importance of vesicles and their trafficking [21], which are particularly important for normal functioning of the immune system. For example, vesicle trafficking is essential for phagocytosis of microbes and later destroying of the microorganisms, antigen presentation, antibody and cytokine secretions, and the movements of immune cells. Therefore, LRBA’s subcellular localization and involvement in the dynamic vesicle trafficking human cells, which have not been published yet, is critical to understanding the subcellular and molecular mechanisms by which LRBA functions and explain that LRBA deficiency causes a multitude of immune disorders and other severe medical conditions.
Monoclonal antibodies against the following proteins were purchased from BD Biosciences (San Jose, California, USA): Caveolin 1, EEA1, GM130, CD49b (Integrin a2), Lamp-1, MAP2B, Bcl-2 (Organelle Sampler Kit, 612740, BD Pharmingen™); PKAC, PKARI, PKARIa, PKARIIa, PKARIIb (PKA Sampler Kit, 611420 BD Transduction Laboratories™); GM130, Golgin-84, GS15, GS27, GS28, p115, p230 trans Golgi, Rab8, Syntaxin 6, Vti1a, Vti1b (Golgi Sampler Kit, 611434, BD Transduction Laboratories™) and Adaptin α, Adaptin β, Adaptin γ, Adaptin δ, Amphiphysin, AP180, Clathrin Heavy Chain, EEA1, Rab4 (Coated Vesicle Sampler Kit, 611424, BD Transduction Laboratories™). The LRBA Prestige antibodies (NBP1-90764) were purchased from Novus Biologicals (Littleton, CO, USA). The Alexa Fluor Conjugated secondary antibodies: anti-mouse IgG-Alexa Fluor® 555, and anti-rabbit IgG-Alexa Fluor® 488 were purchased from Life Technologies (Grand Island, NY, USA). Lipopolysaccharides (LPS) from Escherichia coli 0111:B4 (L2630, SIGMA).
HEK293, RAW264.7 and A549 cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained according to the company’s instructions. These cells were cultured in Dulbecco’s Modified Minimum Essential Medium (DMEM) or RPMI1640 supplemented with 10% FBS and penicillin-streptomycin (5,000 IU/ml penicillin and 5,000 μg/ml streptomycin). The mouse studies have been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of South Florida.
HEK293 cells at concentration of 5x106/ml were placed on glass cover slips in DMEM supplemented with 10% Fetal Calf Serum (FBS) (Gemini Bio-Products, West Sacramento, CA), penicillin and streptomycin. After 24 h, cells were fixed, permeabilized, and stained following the immuno-fluorescence staining protocol from the Human Protein Atlas Project (https://www.sigmaaldrich.com/ life-science/cell-biology/antibodies/prestige-antibodies/prestige-antibodies-in-immunofluorescence-applications.html) [22]. Briefly, growth medium was removed and the cells were washed in 1x PBS, the cells were fixed for 15 minutes in ice cold 4 % paraformaldehyde pH 7.2-7.3 in growth medium supplemented with 10 % FBS. The cells were permeabilized 3 times for 5 minutes each with 0.1 % TRITON® X-100 in PBS. The cells were washed with 1x PBS and incubated overnight at 4 °C with the primary antibodies in 1x PBS supplemented with 4 % FBS. The following day the cells were washed 4 times for 10 minutes each with 1x PBS and incubated for 1.5 hours at room temperature with the secondary antibodies in 1x PBS supplemented with 4% FBS. The cells were counterstained for 4 minutes with the nuclear stain DAPI (0.6 μM in 1x PBS). The cells were washed 4 times for 10 minutes with 1x PBS and then mounted in glycerol + 10 % 10x PBS. The LRBA antibodies and three Golgi protein primary antibodies were used at 1:500 (volume to volume dilution). The anti-mouse IgG-Alexa Fluor® 555 and anti-rabbit IgG-Alexa Fluor® 488 secondary antibodies were used at 1:400.
The confocal imaging was acquired with an Olympus FV1000 MPE multiphoton laser scanning microscope, featuring ultra-high, 2-nanometer (nm) wavelength resolution, using 60x objective (U Plan APO 1.42 N.A. oil) and sequential scanning with 0.5 μm per slice, the use of narrow band emission filters. Single and unlabeled controls were used to assess bleed-through, which produces false co-localization. Co localization analysis for dual stained samples was carried out using JACop and FV10-ASW software (Olympus Corporation, Tokyo, Japan).
RAW264.7 macrophage cells stably transected with pBWEGFP were cultured in a glass bottom dish and put into the mini cell culture chamber at 37 ˚C and 5% CO2 in a Leica TCS SP2 laser scanning inverted confocal microscope. LPS was added at final concentration of 100ng/ml. The time-lapse video of four living cells was taken at one picture per second using the Avg. time lapse interval: 1000.05 ms (1.0 Hz) (+/- 8.9 ms); Initial SAC Position: 1700; Objective: 1 OOx (magnification: 100.00x); Mag. changer: EMCCD (magnification: 1.00x); 8 inning: 1 x 1; Channel 1: c488em at 50 ms. Gain: 3 Intensification: 700 ND: 60 (Independent).
JACoP (Just Another Co localization Plugin) that integrates current global statistic methods and a novel object-based approach [17] and was used for colocolization assay. All of the following features were used: Calculating a set of commonly used co-localization indicators: Pearson’s coefficient, Overlap coefficient, k1&k2 coefficients, Manders’ coefficient, Cytofluorogram for generating commonly used visualizations, Costes’ automatic threshold, Costes’ randomization, Li’s intensity correlation coefficient, and two objects based methods: distances between centers and center-particle coincidence.
The following subcellular colocalization results about LRBA with Golgi proteins, components of coated vesicle proteins and other organelle-specific proteins, and PKA subunits were obtained by immunofluorescence staining and confocal microscopy, and the P-value of 100% for each colocalization was obtained by the Just Another Colocalization Plugin (JACoP) calculation based on the Costes’ randomization colocalization, suggesting that colocalization in the regions masked in white is highly probable [17]. LRBA does not have significant colocalization with other proteins listed in the Materials and Methods section, e.g. PKARI, Bcl-2,MAP2B,Golgin-84, GS15, GS27, Adaptin δ, Amphiphysin, AP180.
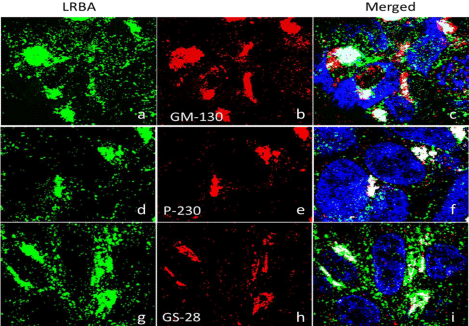
The GC is now viewed as the “headquarters” for signal transduction and cell-fate decisions, controlling mitotic entry, cytoskeleton organization and dynamics, calcium homeostasis, and plasma membrane receptor-initiated and organelle-autochthonous signaling events, in addition to its “post office role” of cargo sorting/processing [20]. The colocalization of LRBA with the GC is demonstrated by immunofluorescence confocal microscopy using antibodies against the endogenous LRBA and three Golgi proteins. The results show that LRBA is colocalized with these Golgi proteins: the cis-Golgi Matrix protein-130 (GM130), P-230 and Golgi Soluble N-ethylmaleimide-sensitive factor Activating protein Receptor (SNARE) protein GS28 (Figure 1). The overlap between LRBA and GM-130 is 62% in one million cells. GM-130 is a coiled-coil protein associated with the Golgi apparatus necessary for tethering events in membrane fusion and has a role in maintaining cis-Golgi structure [23]. P-230 is associated with vesicles budding from the trans-Golgi network [24] and required for the regulated secretion of TNF [25]. GS28 is a 28-kDa membrane protein that plays an essential role in mammalian Endoplasmic Reticulum (ER)–Golgi or intra-Golgi vesicle transport [26], found to protect p53 from degradation [27]. The Golgi colocalization of LRBA was also demonstrated in primary mouse cultured adherent bone marrow cells (Figure 2).
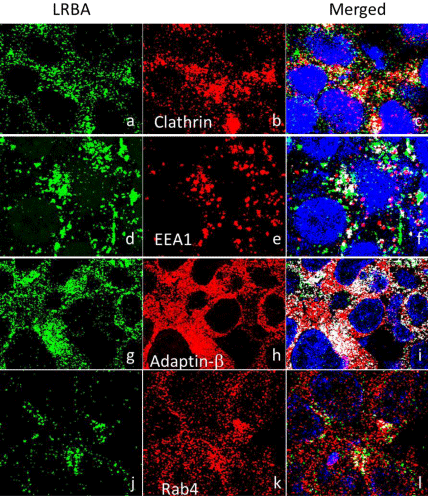
Vesicle trafficking is required for homeostasis (deposition, recycling and degradation) of cell membrane proteins [28]. Previous data show that LRBA regulates two cell membrane receptors: Epidermal Growth Factor Receptor (EGFR) and NOTCH [29-31]. It may also regulate the seven receptors that are associated with CVID [32]. The immunofluorescence confocal microscopy results show that LRBA is colocalized with EEA1, CLATHRIN, RAB4 and ADAPTIN- β(Figure 3). EEA1 is an early endosome marker; Clathrin is a vesicle coating protein, and ADAPTIN-β is an adaptor protein that mediates the formation of vesicles by Clathrin-coated pits, through interaction with membrane-bound receptors. Rab4 a member of the RAB family of RAS-related GTP-binding proteins, important regulators of vesicular transport and are located in specific intracellular compartments. RAB4A is a master regulator of receptor recycling from endocytic compartments to the plasma membrane [33].
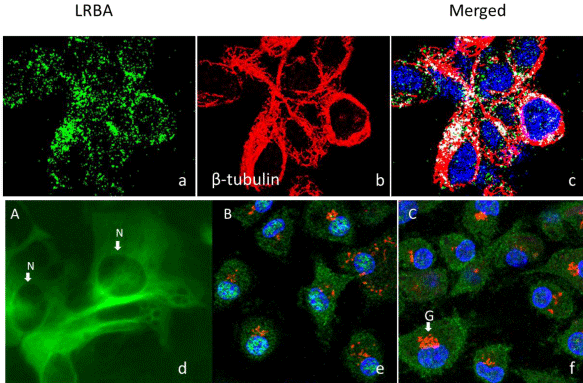
The immunofluorescence confocal microscopy results demonstrate that LRBA in green is colocalized with tubulin in the microtubules in red (Figure 4). The results also were supported by the H293 cells stably transfected with a plasmid that over expresses the full length of Lrba tagged with EGFP at its C-terminal. The cell cytockeleton association of the Lrba/EGFP fusion protein is obvious (Figure 4d).
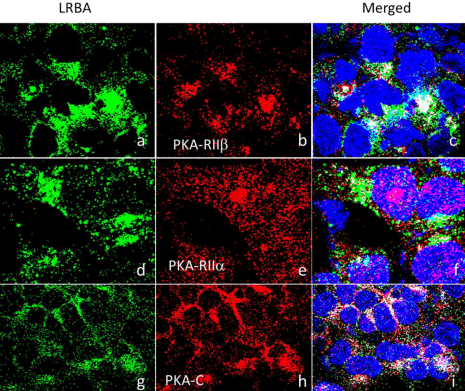
PKA is also known as cAMP-dependent protein kinase. Its function depends on location or its anchor protein and PKA is involved in various membrane trafficking events including the generation of vesicles at the Trans-Golgi Network (TGN) for both constitutive and regulated secretion [30,34]. LRBA is predicted to have two RII binding motifs [8] and its orthologue in Drosophila rugose (rg) is an A Kinase Anchor Protein (AKAP) [16]. LRBA is a potential AKAP and may bind to PKA directly. LRBA is colocalized with the RIIβ, RIIα and RIIc subunits of PKA (Figure 5).
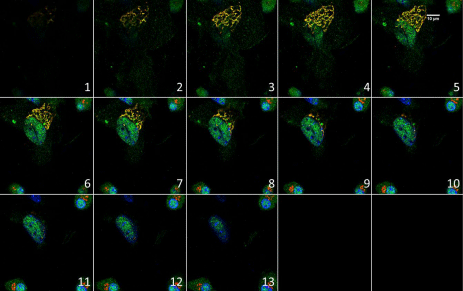
Lrba (gene nomenclature for mouse orthologue of human LRBA) is colocalized with nucleus in mouse bone marrow cells (Figure 2& Figure 4e, f).
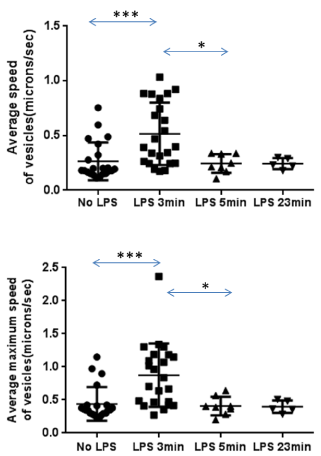
The RAW 264.7 cells were stably transected with a plasmid expressing LRBA BEACH-WD/EGFP fusion protein to study the subcellular localizations of LRBA as described [8]. Confocal time lapse video was used to observe vesicle trafficking (Videos 1 to 6). The results show that LRBA-associated vesicles are isolated from Golgi area, traveling along cytoskeleton, towards cell membrane and fused with the cell membrane. LRBA-associated vesicles forming at the cell membrane were also observed (Video 1& 6). The results confirm the data obtained by immunofluorescence confocal microscopy that LRBA is located at the Golgi, vesicle and cell membrane. LRBA associated vesicles are very active in the pseudopodia (Video 1& average speed of vesicles doubles 3 min after adding LPS into the media, but slows down to original levels after 5 and 23 min (Video 1 - 5, Supplemental Table 1).
LRBA is composed of multiple domains: the Concanavalin A-like (ConA) lectin binding domain [35], VHS [36] [VPS (vacuolar protein sorting)-27, Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) domain and STAM (signal transducing adaptor molecule)], RII binding motifs and WBW super domain. The three-dimensional structure of the WDL-BEACH of LRBA has been determined [37]. The WDL domain structurally looks like a Pleckstrin Homology (PH) domain but cannot bind phospholipids but strongly interacts with the BEACH domain and is also called PH domain [37]. The ConA-like lectin domain was proposed to bind oligosaccharide associated with protein traffic and sorting, especially in relation with the vesicle fusion machinery [35]. VHS domain may be involved in vesicular trafficking by binding sorting receptors that move and transfer cargo between the trans-Golgi network and the endosomal compartment [38], suggesting LRBA may be involved in cargo transportation between the two compartments. LRBA potentially may serve as a scaffold to interact with multiple proteins and is involved in the endomembrane system/vesicle trafficking. However, the function of none of these domains has been determined and the exact function of LRBA is unknown [16]. This subcellular localization study may help to better understand the cellular and molecular mechanisms by which LRBA functions in the cell.
A well-characterized LRBA polyclonal antibody and organelle-specific antibodies were used in the present study to detect the corresponding endogenous proteins and study their co-localizations using confocal imaging and a comprehensive toolbox for subcellular colocalization analysis in a variety of cell-lines/primary cells. The toolbox, JACoP, integrates most currently used global statistic methods for various biological colocalization situations [17]. Although, 0.5 of colocalization coefficient was used as a working threshold for colocalization by some scientists, it cannot be used to determine whether a colocalization is true or not. When the colocalization of two proteins depends on the physiological status of the cells, their colocalization coefficient may vary greatly, from 0 to 1. With the advanced imaging technology, low colocalization with a colocalization coefficient less than 0.5 can be detected as shown in this study. Another concern is that we didn’t include an isotype control for each antibody. However, we used many different antibodies to stain the cells and these antibodies would serve as isotype control for each other. We didn’t notice two different antibodies gave same or similar staining patterns for the data presented here, indicating that these antibodies are specific. The colocalization of LRBA with other proteins presented here is confirmed by the various statistic methods included in the JACoP, and thus is highly possible. In addition to using antibodies, full length or partial LRBA was fused with GFP gene to directly study the subcellular localization in response to LPS stimulation.
These results show that LRBA is extensively associated with the endomembrane system, including the Golgi complex, endosomes, plasma membranes, nucleus, pseudopodia and microtubules, and vesicle trafficking, which is responsive to LPS stimulation. Although co-localization by confocal microscopy cannot prove that LRBA physically binds and interacts with these proteins, these results demonstrate that LRBA does associate with these organelle-specific proteins at the cellular structure levels and suggest that LRBA plays a role in vesicle trafficking and signal transduction essential for the immune system [39]. To prove true interactions with these proteins, mutational analysis or a super resolution microscopy for the protein-protein interaction would be required.
The present study demonstrates that LRBA is mainly localized to the GC demonstrated by the co-localization of LRBA with two golgins (GM-130 and P-230) and another GC protein (GS-28). GS28 is a 28-kDa membrane protein that plays an essential role in mammalian ER (endoplasmic reticulum)–Golgi or intra-Golgi vesicle transport [26] and found to protect p53 from degradation [27]. P-230 is associated with vesicles budding from the trans-Golgi network [24] and is required for the regulated secretion of TNF [25]. GM130 is part of a cis-Golgi matrix and has a role in maintaining cis-Golgi structure [40]. GM130 functions as a tethering factor for pre-Golgi carriers undergoing fusion with cis–Golgi cisternae and regulates the fusion of Golgi cisternae into elongated Golgi ribbons [41]. Unlike the present data that LRBA is associated with both cis-and trans-Golgi, previous data from electron microscopy (EM) show that LRBA is associated with the trans-Golgi but not cis-Golgi [8]. The discrepancy with gold-standard immune EM may have several possibilities, the differences in cell types, physiological status, detect target (endogenous vs. ectopic expression) and techniques. However, these data strongly correlate with previous information [8] and the data from Human Protein Atlas Project [42]. Neurobeachin (Nbea) [29,43], an autism candidate gene, has very similar localizations to that of LRBA and is implicated in post-Golgi membrane traffic and regulatory secretion pathway of large dense-core vesicles containing growth factors and hormones [30,44].
The GC is a central organelle in the endomembrane system. It is critical for signal transduction [20]. For example, internalized LPS and TLR4 are found in the GC [45]. Proto-oncogenes Ras and Src are activated on the GC [46,47]. Membrane trafficking is crucial to the transduction of signaling complexes to the GC and other cellular locations [18]. Thus the endomembrane system plays a critical role in signal transduction. LRBA may be imperative to the GC’s function, as LRBA is highly co-localized with GC proteins and LRBA-deficient B cells show an abnormally high number of GCs [11].
PKA is also known as cAMP-dependent protein kinase, composed of two catalytic subunits and two regulatory subunits. Its function depends on location of its anchor protein. In the binding state, PKA is inactive, but four cAMPs bind to regulatory subunits causing conformation change and release PKA, which becomes active. LRBA contains two potential type II Regulatory subunit (RII) binding motifs to bind the RII subunits of cAMP-dependent protein kinase [8]. RII binding motifs for anchoring PKA through the type II Regulatory subunits (RII) [8]. A computer program, which has over 80 percent accuracy to predict RII binding motifs, predicted one of the two potential RII binding motifs [48]. A previous study suggests that LRBA may not bind the RII subunit [43]. However, by closely reviewing the work, it turns out that the LRBA peptide fragment used in that study does not include the two potential PKA RII binding sites [43,48], which are present in human LRBA and LRBA orthologues in mouse, Drosophila and C. elegans [43]. In contrast, experimental data demonstrate that the LRBA orthologue, rg, and LRBA isoform, Nbea, can bind the RII subunits [16,43]. Therefore whether LRBA is an A Kinase Anchor Protein (AKAP) is unclear. The present data show that LRBA is co-localized to several components of PKA: the catalyst unit, RIIβ and RIIα, supporting that LRBA is an AKAP and potentially can phosphorylate substrate proteins such as NFkB [49], which is central to the immune system. The Golgi complex and TGN are major subcellular locations of RII [50,51].
LRBA may be involved in endocytosis and is colocalized with EEA1, Clathrin and Adaptin-β. Adaptins are proteins that mediate the formation of vesicles by Clathrin-coated pits, through interaction with membrane-bound receptors, which suggest that LRBA is involved in endocytosis. LRBA associated vesicles are very active in the pseudopodia, indicating that LRBA may be involved in forming and growth of pseudopodia membrane. Similarly, the LRBA orthologue in C. elegans, SEL-2, is suggested to be involved in endosomal traffic and perhaps in efficient delivery of cell surface proteins to the lysosome in polarized cells [52], and NBEA, which has 75% of protein homology with LRBA, is implicated in membrane protein traffic and is required for the formation and functioning of central synapses [53].
These involvements of LRBA in vesicle trafficking agree with the previous findings that LRBA plays a role in some important cellular processes that require vesicle trafficking including cell proliferation, apoptosis and autophagy[11, 29, 54, 55]. Its deficiency causes a defect in autophagy [11]. LRBA-deficient lymphocytes undergo apoptosis rather than autophagy [11]. This study did not delineate how LRBA fits into any of the known autophagy pathways. Autophagy is an essential, homeostatic process involving the degradation of a cell’s own components through lysosomal machinery. It plays a fundamental role in the intracellular antigen presentation on MHC class-II molecules [56,57]. Deficiency of LRBA suppresses autophagy by about 50% in B cells, indicating that LRBA is critical for the function of autophagy in human lymphocytes [11]. An LC3 Interaction Region (LIR), which is involved in autophagy, is also predicted in LRBA and it is hypothesize that LRBA is involved in autophagy through association with LC3 [55]. However, confocal microscopy failed to prove that the LIR of LRBA co-localizes with LC3 when autophagy was induced (data not shown). It is possible that the time of the interaction is too short, more sensitive method is required for the detection of the interaction.
In summary, this study provides visual seeing-is-believing evidence at subcellular level that LRBA is extensively associated with the endomembrane/vesicle trafficking system including the Golgi complex, endosomes, plasma membrane, pseudopodia and microtubules. These data suggest that LRBA is an important vesicle trafficking and signal transduction regulator required for the regulation and function of many immune molecules. LRBA deficiency thus may cause defective trafficking and signaling of immune effect or molecules, resulting in immune disorders (immunodeficiency, autoimmunity and inflammation) and other medical conditions manifested in LRBA deficient patients.
This work was supported by the Joy McCann Culverhouse Endowment to the Division of Allergy, Department of Internal Medicine. We would like to acknowledge the USF Health Lisa Muma Weitz Advanced Microscopy & Cell Imaging Core Laboratory under the direction of Dr. B. Jake Cha for assistance in confocal microscopy and imaging analyses.
LRBA is co-localized with Golgi proteins GM-130, P-230 and GS28. Golgi matrix protein GM-130: Pearson’s coefficient=0.562, overlap coefficient =0.674. Trans-Golgi network-localised protein p230: Pearson’s coefficient =0.443, overlap coefficient=0.469. Golgi SNARE GS28: Pearson’s coefficient =0.366, overlap coefficient =0.381.
Lrba is co-localized with Golgi and nucleus in mouse bone marrow cells. The images were acquired with an Olympus FV1000 scanning confocal microscope by sequential scanning with 0.5 μm per slice and labeled sequentially with numbers. Red=Golgi; Green=Lrba. Yellow indicates Lrba colocalized with Golgi.
LRBA is co-localized with Clathrin, early endosome protein EEA1 and Adaptin-β. Clathrin: Pearson’s coefficient=0.363, overlap coefficient=0.403. Early Endosome Antigen 1 (EEA1): Pearson’s coefficient=0.153, overlap coefficient=0.193. Adaptin-β: Pearson’s coefficient=0.234, overlap coefficient=0.345.
LRBA is co-localized with β-tubulin and translocated to the nucleus upon LPS stimulation. Top panel: a. LRBA; b. Tubulin; c. Merged. Pearson’s coefficient=0.171, overlap coefficient=0.207. Bottom panel: d. The plasmid pLrbaEGFP containing full length mouse Lrba coding sequence fused with EGFP at C-terminal was transfected into HEK293 cells with lipofectamine 2000 and selected with G418 at 400μg/ml for 20 days. The LrbaEGFP fusion protein subcellular location pattern has the cytoskeleton pattern. e & f. Mouse (C57BL/6J) bone marrow cells were cultured on glass cover slips with (B) or without (C) LPS (1μg/ml) stimulation for 3 hours, then fixed, permeabilized, and stained. Green=Lrba; Red=GM130; Blue=Nucleus; N=Nucleus; G=Golgi complex.
LRBA is co-localized with the RIIβ, RIIα and PKA Catalytic subunits (PKA-C) of PKA. The endogenous LRBA and the PKA subunits were detected by immunofluorescence confocal microscopy and the results show that LRBA is colocalized with the RIIb and RIIc subunits of PKA. RIIb Pearson’s coefficient=0.225; Overlap coefficient=0.257; RIIa PKA-c Pearson’s coefficient=0.197, overlap coefficient=0.261.
LPS stimulates Lrba-positive vesicles movements. RAW264.7 macrophage cells stably transfected with pBWEGFP were cultured in a glass bottom dish and put into the culture chamber. LPS was added at final concentration of 100ng/ml. The time-lapse video of four living cells was taken at one picture per second using the Leica TCS SP2 laser scanning inverted confocal microscope with a mini cell culture chamber at 37 °C and 5% CO2. Top panel: Average speed of LRBA-associated vesicles; Bottom panel: Average maximum speed of LRBA-associated vesicles; The two tails and unpaired T test p values: ***p<0.001; *p<0.0138 (top panel); ***p<0.0005; *p<0.0117 (bottom panel). LPS 3 min indicates that the measurements were obtained 3 min after adding LPS into the media.
Two dimensional time lapse video of living macrophage cells. RAW264.7 macrophage cells were stably transected with a plasmid expressing LrbaGFP fusion protein and cultured in the mini cell culture chamber in a laser scanning inverted confocal microscope. LPS was added at final concentration of 100ng/ml. The time-lapse video of living cells was taken at one picture per second using the Leica TCS SP2 laser scanning inverted confocal microscope with a mini cell culture chamber at 37 °C and 5% CO2. The GFP fluorescence shows cell membrane, Golgi and vesicle locations of LRBA, and the dynamic activities of LRBA associated vesicles moving from GC to the membrane. Some vesicles were manually labeled as blue spots and numbered in the videos, so the path movement statistics, i.e. Average Speed (microns/sec), Maximum speed (microns/s), Total Displacement (microns), Path Duration (time points), Path Duration (sec), can be calculated. Video 1: No LPS stimulation (Cell#1).
Please download Video here
No LPS stimulation (Cell#2).
Please download Video here
LPS stimulation for 3 minutes.
Please download Video here
LPS stimulation for 5 minutes.
Please download Video here
LPS stimulation for 23 minutes.
Please download Video here
Dynamic movements of LRBA associated vesicles. No LPS stimulation.
Please download Video here