
Research Article
Austin J Clin Immunol. 2023; 9(1): 1050.
Persistent SARS-Cov-2 Infection in an Immunocompromised Host. Role of the Cellular Immune Response
Duarte A1*, Cruces L2,7, Nabaes Jodar MS4,5, Goya S5, Monzani C2,7, Herrera M2,7, Mazzitelli I2,3, Turk G2,3, Guevara D6, Viegas M4,5, Quiroga MF2,3, Laufer N2,3 and Cuesta MC1
1Servicio de Enfermedades Infecciosas Clínica Zabala,Buenos Aires, Argentina.
2CONICET-Instituto de Investigaciones Biomédicas en Retrovirus y SIDA (INBIRS), Universidad de Buenos Aires, Argentina.
3Departamento de Microbiología, Parasitología eInmunología, Facultad de Medicina, Universidad de Buenos Aires, Argentina.
4Consejo Nacional de Investigaciones Científicas y Técnicas, Buenos Aires, Argentina.
5Laboratorio de Virología Hospital de Niños RicardoGutiérrez, Buenos Aires, Argentina.
6Laboratorio de Biología Molecular Swiss Medical Group, Buenos Aires, Argentina.
7Facultad de Medicina, Universidad de Buenos Aires, Argentina.
*Corresponding author: Duarte AServicio de Enfermedades Infecciosas, Clínica Zabala. ZIP Code C1426AAB, Buenos Aires, Argentina.
Received: December 29, 2022; Accepted: February 02, 2023; Published: February 09, 2023
Abstract
We present a patient with B-cell depletion and persistent SARS-CoV-2 infection, for 19 weeks in whom the emergence of viral mutations was documented during its evolution, no humoral response was detected, however a specific cellular response was evidenced, highlighting the role of this in the control of infection.
Keywords: Persistent SARS-CoV-2; Immunocompromised;B-cell depletion; Cellular immune response; Mutations
Introduction
Since the onset of the pandemic, cases of persistent SARS-CoV-2 in immunocompromised patients with higher morbidity and mortality have been reported.
The dynamics of viral shedding, transmissibility and the ability to develop an effective humoral and cellular immune response, particularly given the continuous emergence of new variants and vaccination in these patients, continues to be a subject of research [1].
We present a patient with B-cell depletion and persistence of positive SARS-CoV-2 RT-PCR along 19 weeks despite having received treatment with corticosteroids, Convalescent Plasma (CP) and remdesivir. Direct genome sequences were obtained with a pattern of substitutions characteristic of the B.1.1.7 (alpha) lineage and the presence of mutations during its viral persistence. Although specific humoral response was not detected, there was evidence of a specific cellular response that was enhanced after the first dose of the vaccine.
Case report
70-year-old man with B-cell depletion due to mantle cell lymphoma receiving Rituximab maintenance. He was diagnosed with SARS-CoV-2 infection in April 2021 and was admitted to hospital twice because he developed three symptomatic courses of Covid-19, first mild, then severe, and finally severe-critical pneumonia with periods of improvement in between. Standard of care with oxygen therapy and dexamethasone was administered. In the last episode, he received empiric antibiotics, CP for 2 days and remdesivir for 10 days, showing a rapid clinical-radiological and gasometric response.
On day 66 of diagnosis, he was discharged. In outpatient controls for 19 weeks, persistence of detectable SARS-CoV-2 RT-PCR was observed with all Ct ≤ 26. The first negative RT-PCR was obtained on day 148 (Figure 1A). Two months later, he received the first dose of vaccine for Covid-19.
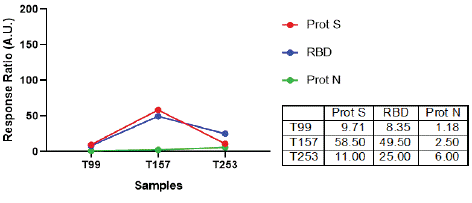
Figure 1A: Timeline of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)-related investigations. Ct, cycle threshold; orf 1ab: gen ORF 1ab; N, nucleocapsid; T: time; IgM/IgG: immunoglobulin M/G; PCR: Real time polymerase chain reaction; NPS: nasopharyngeal.
Additional and complementary assays were performed. Evaluation of SARS-CoV-2 RNA in plasma was performed on day 49 after symptom onset with a non-detectable result. In viral infectivity studies, cytopathic effect on Vero cells was not observed but SARS-CoV-2 RNA was detected in the cell culture supernatant. Direct viral sequences were obtained from 7 Nasopharyngeal Samples (NPS) for the region comprising amino-acids 340 to 574 of the S protein. All sequences presented the pattern of substitutions characteristic of the B.1.1.7 (alpha) lineage, with only 2 non-synonymous substitutions with respect to the reference sequence (GISAID Accession ID EPI_ISL_ 601443). In order to analyze the intra-host evolution throughout the persistent infection, NGS sequencing by Illumina platform was performed in four NPS with adequate viral RNA in quantity and quality at different time points of infection (40, 78, 98 and 125 days after the first positive PCR: T40, T78, T98 and T125, respectively). The analysis of the Variant Call Format (VCF) files showed a clear intrahost variation across the four analyzed samples at the amino acidic level (mainly in S and nsp3 proteins) (supplementary table A). At the sample level, mixed viral populations were found. The most interesting result was found in T78, which was taken after convalescent plasma therapy 56 days after diagnosis with a mixed population in four positions of the N Terminal Domain (NTD) of the S protein (Supplementary Figure 1 A). The detailed results of the NGS analyses of the four samples are described in Supplementary material.
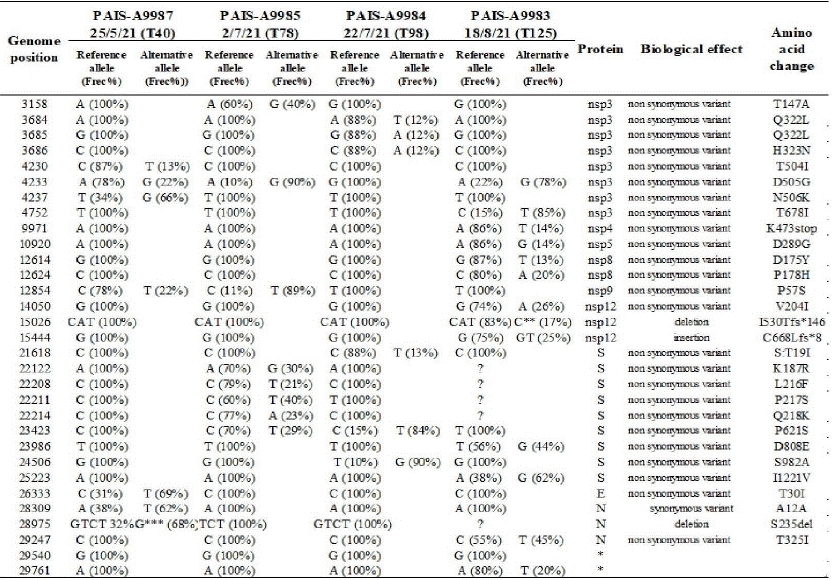
Table A. Overview of the SARS-CoV-2 variant allele frequencies across four samples. The table summarizes the positions in the viral genome and the variant frequencies in the different samples using a cut off value of 5%. The reference allele corresponds to the hCoV-19/Wuhan/WIV04/2019 genomic sequence (EPI_ISL_402124) and variant frequencies are indicated in parentheses. The symbol ? represents low coverage sites. In addition, the locations of deletions, insertions and synonymous and nonsynonymous mutations with their respective amino acid changes are shown.
Assays to detect IgG against Spike (COVIDAR) and against nucleocapsid (N, CMIA) proteins were performed on several samples, including those obtained after vaccination (Figure 1A) but all showed negative results. However, specific cellular responses against N and S proteins were evidenced. Cellular immune response was evaluated at months 2 and 3 during infection and also 3 months after the last positive PCR result. For that, ELISpot was performed to detect Interferon-gamma producing cells after stimulation with SARS-CoV-2 Wuhan variant peptide pools and proteins. More specifically, PBMCs were stimulated in the presence of Spike, RBD and Nucleocapsid proteins or peptide pools from S and N viral proteins (co-stimulation with anti-human CD28 and CD49d was performed).
A gradual increase in the response against Spike and Nucleocapsid peptide pools was observed, reaching the highest values after the PCR negative result. In particular, Spike-specific responses were markedly higher compared to N-specific responses as time progressed, i.e., month 3 after infection and month 3 after the first negative SARS-CoV-2 PCR result (Figure 1B). No

Figure 1B: Cellular immune response after in vitro stimulation of PBMCs with SARS-CoV-2 peptide pools. IFN-γ ELISpot assays were performed to determine the frequency of Ag-experienced T cells in peripheral blood at the indicated times (T99, T157 and T253). Stimulation of BPMCs with peptide pools encompassing S protein or N protein was performed. Afterwards, IFN-γ producing cells were determined as illustrated in the Supplementary materials section.
Discussion
Given that the dynamics of SARS CoV-2 is the result of a complex balance between its replication, the immune response and possible therapeutic options, it is expected that immunosuppressed patients may offer the virus better opportunities to persist.
The humoral immunity deficit of our patient due to his underlying disease and the anti-tumor treatment he received explains the absence of seroconversion detected even after vaccination, as well as the difficulty to eliminate the virus [1].
Coronaviruses accumulate mutations at a much slower rate than other RNA viruses, and most of these have little or no phenotypic impact. However, some can significantly influence viral transmissibility and the ability of the virus to escape the host’s immune response [2-4].
Whole genome sequencing of SARS-CoV-2 in persistent infections is key to understanding and expanding knowledge about intra-host evolution and variant acquisition (o diversity acquisition). These modifications could impact the ability to confer resistance to neutralizing antibodies leading to partial immune escape or otherwise compensate for infectivity deficits associated with other non-synonymous substitutions.
The alpha variant (B.1.1.7), one of the first emerging lineages carrying S: N501Y, spread in southeastern England in early 2020 and quickly became dominant in the world. Despite being significantly more transmissible than wild-type genotypes, Alpha was not associated with significant immune escape from the neutralizing activity of convalescent or vaccinated sera [3,4].
In our patient, the study of the genome on 7 samples showed the presence of the successful variant B.1.1.7, with the presence of intravariant mutations. Viral infectivity was demonstrated by the presence of SARS CoV-2 RNA in the viral culture supernatant, despite the absence of cytopathic effect.
This case highlights a still unresolved problem of immunocompromised hosts, which can spread infectious virus for longer periods of time and, since it is not possible to carry out routine viral cultures to demonstrate their infectivity, alternative criteria are required to define isolation times. The problem becomes even more complex when considering that they are also a permanent source of emergence of variants and therefore perpetuation of the ongoing pandemic with implications for the effectiveness of currently available therapeutics and vaccines.
Several studies provide data on the virus and its evolutionary dynamics, on humoral immunity but the role of the cellular response in these patients is less well understood. Trials that analyze the responses of these patients to vaccines maintain that when they are evaluated through the combination of serology and cell-based assays, a lower response is observed compared to healthy individuals [1-4].
Patients with hematologic malignancies show complex and escalating immunologic consequences of SARS-CoV-2, including T-cell depletion, malignant B-cell vulnerability and impaired seroconversion with subsequent chronic or recurrent COVID disease. Experience with the SARS-CoV-2 treatment in this group is still scarce [5].
Remdesivir aims to reduce viral load before the virus triggers a powerfully devastating hyper inflammatory response. In immunocompetent patients it is administered within the first 7-10 days after the onset of symptoms, since viral replication spontaneously decreases thereafter, and subsequent therapy does not prevent the possible cytokine storm [6,7]. Remdesivir monotherapy is associated with treatment failure, but combination with CP appears to be a promising therapy [8-10].
This case indicates that the simultaneous use of remdesivir and CP for protracted COVID-19 in B-cell deficient patients could lead to viral clearance when antigen-specific T cells are elicited. These results also can argue against the concept that the beneficial effect of remdesivir and CP is achieved exclusively when they are administered at the beginning of the infection [11,12].
We believe that this case report reinforces the importance of antigen-specific T-cell response for the virus control and impact on clinical outcome. A more thoughtful knowledge about the SARS CoV-2 specific cell response and mechanism to enhance it may contribute to new strategies for monitoring and clinical decision-making in high-risk patients with COVID-19. In conclusion, our results highlight the importance of the immunocompromised patient as a model of viral persistence and source of variants and therefore epidemiological concern about the implications regarding the perpetuation of the pandemic and loss of therapeutic strategies and efficacy of available vaccines.
We also highlight the importance of the different pillars of the immune response in these patients, which are decisive for controlling and limiting the dynamics of the viral infection.
Supplementary materials
SARS-CoV-2 specific cellular responses. PBMC were plated on sterile 96-well plates (MultiScreen IP plates; Millipore), coated with mouse anti-human IFN-γ monoclonal antibody (BD Biosciences) at 2x105 cells/well. SARS-CoV-2 Spike, RBD, Nucleocapside proteins (kindly provided by Dr. A. Gamarnik, Leloir Institute, Buenos Aires, Argentina), Spike or Nucelocapside peptide pools (BEI Resources, NIAID, NIH. NR-52402, and NR-52404) were titrated beforehand and added in duplicate wells (final concentration 10μg/ml for proteins and 2μg/ml for peptide pools). Negative (medium plus 0.05% DMSO) and positive (PHA 10μg/ml Sigma-Aldrich) controls were included for each donor. Plates were developed using biotinylated anti-human IFN-γ monoclonal antibody, streptavidin-peroxidase complex, and AEC (3-amino-9-ethylcarbazole) substrate reagent set (BD Biosciences). Plates were scanned on an ImmunoSpot reader and specific spots were counted using the ImmunoSpot software (Cellular Technology Ltd.).
Acknowledgements
We gratefully acknowledge Dr. Nicolas Bobrovsky for his contribution in the outpatient follow-up of the patient.
Funding statement: This study received no funding or donations.
Conflict of interest: None declared.
References
- Haidar G, Mellors JW. Improving the Outcomes of Immunocompromised Patients With Coronavirus Disease 2019. Clin Infect Dis. 2021; 73: e1397-e1401.
- Boehm E, Kronig I, Neher RA, Eckerle I, Vetter P, et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021; 27: 1109-1117.
- Gerdol M, Dishnica K, Giorgetti A. Emergence of a recurrent insertion in the N-terminal domain of the SARS-CoV-2 spike glycoprotein. Virus Res. 2022; 310: 198674.
- García-Vidal C, Iglesias-Caballero M, Puerta-Alcalde P, Mas V, Cuesta-Chasco G, et al. Emergence of Progressive Mutations in SARS-CoV-2 From a Hematologic Patient With Prolonged Viral Replication. Front Microbiol. 2022; 13: 826883.
- Cesaro S, Ljungman P, Mikulska M, Hirsch H, Lilienfeld-Toal M, et al. Recommendations for the management of COVID-19 in patients with haematological malignancies or haematopoietic cell transplantation, from the 2021 European Conference on Infections in Leukaemia (ECIL 9). Leukemia. 2022 Jun;36(6):1467-1480.
- Abdul-Jawad S, Baù L, Alaguthurai T, Del Barrio I, Laing A, et al. Acute Immune Signatures and Their Legacies in Severe Acute Respiratory Syndrome Coronavirus-2 Infected Cancer Patients. Cancer Cell. 2021; 39: 257-275.e6.
- Brown LK, Moran E, Goodman A, Baxendale H, Bermingham W, et al. Treatment of chronic or relapsing COVID-19 in immunodeficiency. J Allergy Clin Immunol. 2022; 149: 557-561.e1.
- Beigel JH, Tomashek KM, Dodd LE, Mehta A, Zingman B, et al. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med. 2020; 383: 1813-1826.
- Prendecki M, Clarke C, Edwards H, McIntyre S, Mortimer P, et al. Humoral and T-cell responses to SARS-CoV-2 vaccination in patients receiving immunosuppression. Ann Rheum Dis. 2021; 80:1322-1329.
- Mrak D, Tobudic S, Koblischke M, Graninger M, Radner H, et al. SARS-CoV-2 vaccination in rituximab-treated patients: B cells promote humoral immune responses in the presence of T-cell-mediated immunity. Ann Rheum Dis. 2021; 80: 1345-1350.
- Jassem J, Marek-Trzonkowska NM, Smiatacz T, Arcimowicz L, Papak I, et al. Successful Treatment of Persistent SARS-CoV-2 Infection in a B-Cell Depleted Patient with Activated Cytotoxic T and NK Cells: A Case Report. Int J Mol Sci. 2021; 22: 10934.
- D’Abramo A, Vita S, Maffongelli G, Beccacece A, Agrati Ch, et al. Clinical Management of Patients With B-Cell Depletion Agents to Treat or Prevent Prolonged and Severe SARS-COV-2 Infection: Defining a Treatment Pathway. Front Immunol. 2022; 13: 911339.