Abstract
The species validity of the genus Spirometra tapeworm is still controversial. Here, we aimed to investigate molecular characteristics with a total of 11 Spirometra samples collected from Asia and USA. They were all examined based on mitochondrial cytochrome c oxidase subunit 1 (CO1) and NADH dehydrogenase subunit 1 (ND 1) gene sequences. The CO1 sequences of 7 Spirometra samples from Korea (n=3), China (n=2), and USA (n=2) were presumptively designated into S. mansoni (99.1-99.9% identities), but they were hardly differentiated to S. decipiens (99.0-99.8% identities) with the only one base pair-difference. The ND1 sequences of above 7 samples showed 100% identity with S. decipiens, but we could not compare them with those of S. mansoni due to lacking database. The CO1 sequences obtained from 3 Myanmar and 1 Cambodia samples were supposed to be new Spirometra species. The ND1 sequences of 2 Myanmar samples revealed 100% identity with S. ranarum (SP1803) and S. decipiens (SP1804), respectively. Seven samples of S. mansoni from Korea, China and USA were co-clustered without the exact differentiation among S. decipiens or S. erinaceieuropaei. Three Myanmar samples were relatively close to each other, but quite different to a Cambodian Spirometra species. We found that several CO1 sequences of S. mansoni, S. decipiens, S. erinaceieuropaei deposited in GenBank were quite similar. Our data showed that it is not easy to differentiate between S. mansoni, S. decipiens, and S. erinaceieuropaei even with CO1 sequences and the expansion of ND1 sequences may be further needed for their differentiation.
Keywords: Spirometra, molecular characteristics; Mitochondrial cytochrome c oxidase subunit 1; NADH dehydrogenase subunit 1, Asia, USA
Introduction
Genus Spirometra species (Cestoda: Diphyllobothriidae) tapeworms mainly inhabit in the small intestines of canine and feline hosts worldwide and their plerocercoid larva, sparganum (pl. spargana), causes sparganosis in humans [1]. Humans are accidental hosts and acquire the parasite by ingesting water contaminated with copepods carrying procercoids, by ingesting uncooked or undercooked meat of the second intermediate and/or paratenic hosts with plerocercoids, or by using the frog and snake flesh as a poultice in traditional medicine [2]. In humans, the ingested sparganum via the gastrointestinal track usually migrates to subcutaneous tissues and muscles of the abdominal wall and limbs, where it forms a nodule and presents as a creeping eruption [3]. Spirometra spp. tapeworms are found worldwide but human infections with larval and adult worms are most often reported in Asian countries, typically China, Republic of Korea (Korea), Japan and Thailand [4-6]. Until now, nearly 64 nominal species have been described in the genus Spirometra, but only 4 are considered valid [7-9]. The life cycle of Spirometra species have been studied and their genus name were validated by several researchers. Joyeux and Houdemer (1928) infected dogs with spargana obtained from various animals in India and recovered specimens, naming them Diphyllobothrium mansoni [10]. Faust et al. (1929) infected dogs and cats with spargana detected in Chinese hedgehogs (Erinaceus dealbatus) and named the recovered specimens as Diphyllobothrium erinacei [11]. On the other hand, Iwata (1934) referred to the species distributed in the Far East as D. erinacei, Joyeux et al. (1934) named those found in Southeast Asia as D. mansoni [12], Mueller (1935) named those distributed in North America as Diphyllobothrium mansonoides, and Brumpt (1936) listed those found in Europe as D. erinaceieuropaei [13, 14]. In Korea, Lee et al. (1990) conducted laboratory experiments to complete the life cycle of Spirometra species distributed in Korea and based on their biological and morphological characteristics, they named the species distributed in Korea as S. erinacei [15].
Adult worms of Spirometra species is traditionally identified by examination of the reproductive system, especially the number and shape of uterine coils, but it is not enough to distinguish some of Spirometra based on morphological characteristics only. The word “sparganum” is a generic name for a plerocercoid, a tissue-migrating larva of tapeworms of the Diphyllobothrium and Spirometra species, when their adult worm is unknown [4]. The recent progress in genetic evidence by DNA barcoding methods, along with the analysis of morphological features, allows for the support of the distinctiveness of Spirometra species [9]. Recently, mitochondrial genome sequences from spargana have been extensively reported and mitochondrial genome sequences of Spirometra erinaceieuropaei, Spirometra decipiens and Spirometra ranarum have been addressed [16-18]. Some researchers reported that S. decipiens, S. ranarum and S. mansoni might be regarded as synonyms of S. erinaceieuropaei, however this remains to be verified also [19]. Herein, we investigate molecular characteristics based on mitochondrial cytochrome c oxidase subunit 1 (CO1) and NADH dehydrogenase subunit 1 (ND 1) gene sequences among several spargana specimens collected from several Asian countries and USA.
A total of 11 samples, including 10 spargana and 1 adult Spirometra, were collected in the naturally infected snakes and frogs, and experimentally infected tadpoles, mice and cat between 2002 to 2019 (Supplemental Table 1). All specimens were kept in -80 deepfreezer or were preserved in 70% ethanol for experimental use. Total genomic DNA was extracted from each samples using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol, and used as a template for PCR. The two kind of primer sets were used; Spi-CO1F (5'- GACTAAGTGTTTTCAAAACACTAAGTG -3') and Spi-CO1R (5'- CACCCTACCCCTGATTTACAAAAT -3') for CO1 gene, and Spi-ND1F (5´- GGAGAATATTGGTTTGTCTAACCA -3´) and Spi-ND1R (5´- CCTTCTTAACGTTAACAGCATTACGAT -3´) for ND1 gene [16]. The PCR products were sent to Macrogen (Seoul, Korea) for direct sequencing using the PCR primers listed above. The chromatograms of both sequences were trimmed manually and assembled using SeqMan software (DNASTAR, Madison, Wisconsin, USA). The assembled sequences were compared with Spirometra species sequences deposited in GenBank using blast searches. The phylogenetic relationships among the sequences were inferred using neighbor-joining (NJ) analysis with Geneious software (11.1.3) based on Tamura-Nei genetic distance model. Bootstrap analysis was performed with 1,000 replication. The CO1 sequences of the Spirometra samples were compared with the reference CO1 sequences of S. mansoni, S. decipiens, S. erinaceieuropaei, and S. ranarum deposited in GenBank (accession number LC328896, MW031094, KX528074, MH298843) summarized in Table 1. The CO1 sequences obtained from 3 Korean isolates (SP0301, SP0501, SP1801) of Spirometra species ranging 1,521-1,537 bp showed 99.9% identity with S. mansoni and 99.8% identity with S. decipiens, but 99.6% identity with S. erinaceieuropaei. The CO1 sequences obtained from 2 Chinese samples (SP0201, SP0401) ranging 1,523-1,531 bp showed 99.1-99.9% identity with S. mansoni and 99.0-99.8% identity with S. decipiens, but 98.8%-99.6% identity with S. erinaceieuropaei. The CO1 sequences of 2 samples (SP0202, SP0203) obtained from USA ranging 1,523-1,555 bp showed 99.4-99.9% identity with S. mansoni and 99.4-99.8% identity with S. decipiens, but 98.1%-99.6% identity with S. erinaceieuropaei. Overall, above 7 Spirometra species samples from Korea, China and USA were presumptively designated into S. mansoni, although they were hardly differentiated to S. decipiens with the only one base pair-difference. The CO1 sequences obtained from 3 Myanmar samples (SP1802, SP1803, SP1804) ranging 1,503-1,564 bp showed 98.1-98.9% identity with S. mansoni and 98.0-98.8% identity with S. decipiens, 97.7%-98.6% identity with S. erinaceieuropaei, and 97.8-98.2% identity with S. ranarum, respectively. A Cambodian sample (SP1901) with 1,536 bp showed 98.6% identity with S. ranarum, but 97.2% identity with S. mansoni, 97.1% identity with S. decipiens, and 96.9% identity with S. erinaceieuropaei, respectively. Overall, the exact species level could not be designated to above 4 samples from Myanmar and Cambodia by CO1 sequences, suggesting that they were supposed to be new Spirometra species. The ND1 sequences of the 11 samples of Spirometra species were compared with the reference ND1 sequences of S. decipiens, S. erinaceieuropaei, and S. ranarum which were deposited in GenBank (accession number MW031094, MN432159 and MH298844). We could not compare with the ND1 sequences of S. mansoni, due to lacking enrolled ND1 sequences of S. mansoni deposited in GenBank yet. The ND1 sequences of 8 samples from Korea (n=3), China (n=2), USA (n=2), and Myanmar (n=1) showed 100% identity with S. decipiens, but 97.7% identity with S. erinaceieuropaei or S. ranarum. One Spirometra sample from Myanmar (SP1803) showed 100% identity with S. ranarum, but 97.9% identity with S. decipiens and 98.7% identity with S. erinaceieuropaei. ACambodian sample (SP1901) showed 98.2%, 98.9%, and 98.5% identities with S. decipiens, S. erinaceieuropaei, and S. ranarum, respectively. The phylogenic tree showed that 7 Spirometra samples from Korea, China and USA presumptively designated into S. mansoni were co-clustered without the exact differentiation among S. mansoni, S. decipiens, and S. erinaceieuropaei (Figure 1). Three Myanmar samples were relatively close to each other, but quite different to a Cambodian Spirometra species. We found that several CO1 sequences of S. decipiens, S. erinaceieuropaei deposited in GenBank were quiet similar and they could not well differentiated each other.
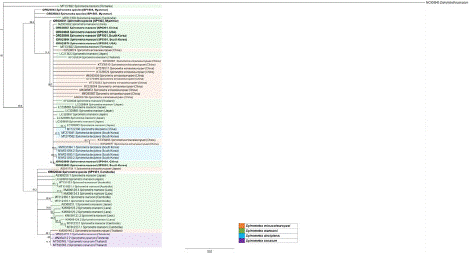
Figure 1: Phylogenic tree constructed based on CO1 sequences from Spirometra samples collected in this study and several Spirometra species from sequences deposited in GenBank. The bootstrap values for the clade with consensus support (%) were displayed on the tree to the left of the most recent common ancestral node for that clade. The scale bar at the bottom of the tree indicated the length of the branches of the tree.
Sample
Identity to the CO1 sequences of various Spirometra species (Accession no./Country)
Identity to the ND1 sequences of various Spirometra species (Accession no./Country)
Final IDa
Accession No.
S.mansoni
S.decipiens
S.erinaceieuropaei
S.ranarum
S.decipiens
S.erinaceieuropaei
S.ranarum
(LC328896/Japan)
(MW031094/ Korea)
(KX528074/China)
(MH298843/Myanmar)
(MT274581/ Korea)
(MN432159/China)
(MH298844/Myanmar)
SP0301
1520/1521
(99.9%)1519/1521
(99.8%)1515/1521
(99.6%)-
878/878
(100%)623/636
(97.9%)860/878
(97.9%)S.mansoni
OR028864
SP0501
1530/1531
(99.9%)1529/1531
(99.8%)1525/1531
(99.6%)-
891/891
(100%)623/636
(97.9%)873/891
(97.9%)S.mansoni
OR028865
SP1801
1536/1537
(99.9%)1535/1537
(99.8%)1531/1537
(99.6%)-
891/891
(100%)623/636
(97.9%)873/891
(97.9%)S.mansoni
OR028866
SP0201
1510/1523
(99.1%)1509/1523
(99.0%)1506/1523
(98.8%)-
891/891
(100%)623/636
(97.9%)873/891
(97.9%)S.mansoni
OR028867
SP0401
1530/1531
(99.9%)1529/1531
(99.8%)1525/1531
(99.6%)-
891/891
(100%)623/636
(97.9%)873/891
(97.9%)S.mansoni
OR028868
SP0202
1515/1523
(99.4%)1514/1523
(99.4%)1510/1523
(99.1%)-
886/886
(100%)623/636
(97.9%)868/886
(97.9%)S.mansoni
OR028869
SP0203
1554/1555
(99.9%)1553/1555
(99.8%)1549/1555
(99.6%)-
891/891
(100%)623/636
(97.9%)873/891
(97.9%)S.mansoni
OR028870
SP1802
1514/1530
(98.9%)1513/1530
(98.8%)1509/1530
(98.6%)1497/1530
(97.8%)-
-
-
Spirometra species
OR026041
SP1803
1536/1564
(98.2%)1535/1564
(98.1%)1531/1564
(97.8%)1537/1564
(98.2%)868/886
(97.9%)628/636
(98.7%)886/886
(100%)Spirometra species
OR026042
SP1804
1503/1532
(98.1%)1502/1532
(98.0%)1498/1532
(97.7%)1499/1532
(97.8%)891/891
(100%)623/636
(97.9%)858/876
(97.9%)Spirometra species
OR026043
SP1901
1493/1536
(97.2%)1492/1536
(97.1%)1489/1536
(96.9%)1515/1536
(98.6%)858/873
(98.2%)629/636
(98.9%)860/873
(98.5%)Spirometra species
OR026044
aIdentification.
Table 1: Matched base pairs (%) of the CO1 and ND1 sequences in 11 Spirometra samples obtained from Asia and USA in this study.
Yamasaki et al. performed haplotype analysis based on mitochondrial CO1 gene sequences and noticed two distinct Spirometra species, Type I and Type II, were present in Asia and neither of which is close to likely European species, S. erinaceieuropaei [3]. In addition, they have pointed out that 2 species, S. decipiens and S. ranarum reported in Asia were conspecific with Type I, and Type I was probably conspecific with S. mansoni, and undescribed Type II species. Similar with this notion, our study also showed that all Spirometra species collected from Korea, China, and USA were the mostly close to S. mansoni. Furthermore, 3 Myanmar and a Cambodian Spirometra species may be rather new ones, relatively close to S. ranarum. In spite of that, we could not completely determine their species names without proper matching results at Genbank database. We found that several CO1 sequences of S. mansoni, S. decipiens, S. erinaceieuropaei deposited in GenBank were quite similar and they could not well differentiate each other. Despite notable attempts to clarify the taxonomy, host specificity, and geographical distribution of Spirometra, it remains still obscure. Collectively, our data showed that it is not easy to differentiate between S. mansoni, S. decipiens, and S. erinaceieuropaei even with CO1 sequences and the expansion of ND1 sequences may be further needed for the species identification of Spirometra species tapeworms.
Molecular identification has played an important role in improving understanding of phylogenetic relationships, genetic variation and taxonomy. With uncertainty in morphology, however, unlabeled or mislabeled molecular data from several cases of sparganosis leaded to many misidentifications and further confusion [8]. It may be not easy to completely differentiate them even with molecular evidence. Several factors such as the quality and quantity (reading length) of sequences, the quality of database, and mitochondrial DNA sequence variation of Spirometra species which might be existed even in the same Spirometra spp. should be considered in their interpretation. Previously, partial mitochondrial DNA sequence variations of S. erinaceieuropaei isolates from China, Japan and Indonesia ranged from 0.0-8.4% for CO1 [20]. According to the latest taxonomy, members of Spirometra have been classified into at least 6 separate species, S. erinaceieuropaei, S. folium, S. mansoni, Spirometra sp. 1, and the tentative assignment of S. decipiens complex 1 and 2 [9]. This work described the molecular characteristics based on CO1 and ND 1 gene sequences among several spargana specimens collected from several Asian countries and USA. Our data support above taxonomy with notion that several Korean isolates previously reported as “S. decipiens” might be rather conspecific with S. mansoni. It should be cautious to read CO1 sequences of Spirometra spp. considering that S. mansoni, S. decipiens, S. erinaceieuropaei deposited in GenBank might be quite similar, harboring less discriminative power.
Author Statements
Declaration of Competing Interest
The authors declare no conflicts of interest related to this study.
Data Availability
No data was used for the research described in the article.
Funding
This work was supported by Young Medical Scientist Research Grant through the Daewoong Foundation (DFY2216P) and the Basic Science Research Program through the National Research Foundation of South Korea funded by the Ministry of Education (grant no. NRF- 2022R1C1C1002741).
Author Contributions
Conceptualization: Sohn WM, Won EJ; Data curation: Lee YJ; Formal analysis: Lee YJ; Funding acquisition: Won EJ; Investigation: Lee YJ, Won EJ; Methodology: Lee YJ, Won EJ; Project administration: Won EJ; Supervision: Sohn WM; Writing – original draft: Lee YJ, Won EJ; Writing – review & editing: Lee YJ, Won EJ, Sohn WM.
References
- Liu Q, Li MW, Wang ZW, Zhao GH, Zhu XQ. Human sparganosis, a neglected food borne zoonosis, Lancet Infect Dis. 2015; 15: 1226-1235.
- Cho SY, Bae JH, Seo BS. Some Aspects of Human Sparganosis in Korea, Kisaengchunghak Chapchi. 1975; 13: 60-77.
- Yamasaki H, Sanpool O, Rodpai R, Sadaow L, Laummaunwai P, Un M, et al. Spirometra species from Asia: Genetic diversity and taxonomic challenges, Parasitol Int. 2021; 80: 102181.
- Anantaphruti MT, Nawa Y, Vanvanitchai Y. Human sparganosis in Thailand: an overview. Acta Trop. 2011; 118: 171-6.
- Kim JG, Ahn CS, Sohn WM, Nawa Y, Kong Y. Human Sparganosis in Korea. J Korean Med Sci. 2018; 33: e273.
- Li MW, Song HW, Li C, Xie WT, Lin RQ, Zhu XQ. Sparganosis in mainland China, Int J Infect Dis. 2011; 15: e154-6.
- Fredes F, Mercado R, Salas IP, Sugiyama H, Kobayashi H, Yamasaki H. Morphological observation and molecular phylogeny of Spirometra decipiens complex 1 (Cestoda: Diphyllobothriidae) found in cat from Chile. Parasitol Int. 2022; 87: 102493.
- Scholz T, Kuchta R, Brabec J. Broad tapeworms (Diphyllobothriidae), parasites of wildlife and humans: Recent progress and future challenges, Int J Parasitol Parasites Wildl. 2019; 9: 359-369.
- Kuchta R, Kolodziej-Sobocinska M, Brabec J, Mlocicki D, Salamatin S, Scholz T. Sparganosis (Spirometra) in Europe in the Molecular Era, Clin Infect Dis. 2021; 72: 882-890.
- Joyeux CH, Houdemer E. Recherches sur la faune helminthologique de l’Indochine (Cestodes et Trématodes) (Suite et fin). Ann Parasitol Hum Comp. 1928; 6: 27-58.
- Faust EC, Campbell HE, Kelogg CR. Morphological and biological studies on the species of Diphyllobothrium in China. American Journal of Epidemiology. 1929; 9: 560-583.
- Joyeux TDJ. C, Note sur les cestodes d’oiseaux recoltes dans la region de Marseille., Ann Mus Hist Nat Marseille. 1934; 26.
- Mueller JF. A Diphyllobothrium from Cats and Dogs in the Syracuse Region. The Journal of Parasitology. 1935; 21: 114-121.
- E.m. Brumpt, Pre cis de parasitologie, 5. e d., entierement remanie e, ed., Masson et cie, Paris. 1936.
- Lee SH, We JS, Sohn WM, Hong ST, Chai JY. Experimental life history of Spirometra erinacei, Kisaengchunghak Chapchi. 1990; 28: 161-73.
- Jeon HK, Park H, Lee D, Choe S, Kang Y, Bia MM, et al. Genetic and Morphologic Identification of Spirometra ranarum in Myanmar. Korean J Parasitol. 2018; 56: 275-280.
- Zhang X, Duan JY, Shi YL, Jiang P, Zeng J, Wang ZW, et al. Comparative mitochondrial genomics among Spirometra (Cestoda: Diphyllobothriidae) and the molecular phylogeny of related tapeworms, Mol Phylogenet Evol. 2017; 117: 75-82.
- Zhang X, Wang H, Cui J, Jiang P, Fu GM, Zhong K, et al. Characterisation of the relationship between Spirometra erinaceieuropaei and Diphyllobothrium species using complete cytb and cox1 genes, Infect Genet Evol. 2015; 35: 1-8.
- Waeschenbach A, Brabec J, Scholz T, Littlewood DTJ, Kuchta R. The catholic taste of broad tapeworms - multiple routes to human infection. Int J Parasitol. 2017; 47: 831-843.
- Zhang X, Cui J, Wei T, Li LY, Jiang J, Lu JC, et al. Wang, Survey and genetic variation of Spirometra erinaceieuropaei sparganum in frogs and snakes from Guangxi of southern China. Trop Biomed. 2014; 31: 862-70.
