
Original Article
Austin J Clin Neurol 2022; 9(1): 1159.
Clinical Features and Differential Gene Screening of Invasive Behaviors between Glioma and Brain Metastasis
Zeng X¹, Chen G¹, Dong M², Feng L², Chu L² and Liu J¹*
1Guizhou Medical University, Guiyang, Guizhou, People’s Republic of China
2Department of Neurosurgery, The Affiliated Hospital of Guizhou Medical University, Guiyang, Guizhou, People’s Republic of China
*Corresponding author: Jian Liu, Guizhou Medical University, Guiyang, Guizhou550004, China
Received: October 22, 2022; Accepted: November 18, 2022; Published: November 25, 2022
Abstract
Objective: To analyze invasive behaviors between glioma and Brain Metastasis (BM), and to screen invasive differentially expressed genes.
Methods: Patients diagnosed pathologically with glioma or BM divided into low-grade glioma groups (n = 19), high-grade glioma group (n = 18), and BM group (n = 15). The survival period was determined. The clinical characteristics were retrospectively analyzed to draw Kaplan-Meier survival curve. Glioma and BM samples were obtained for RNA sequencing. By GO, KEGG, PubMed, and GeneCard, invasive genes were theoretical selected. Correlation between invasive genes and pathological grade was performed. The expression level of invasive genes was verified.
Results: The survival curve found that the clinical invasive behavior related to short lifetime includes: size, edema range, blood supply, vascular invasion, grade, and Ki 67 expression. Theoretical analysis finally found that the expression levels of CALM3, CAMK2A, CAMK2B, and PRKCG were negatively correlated with pathological grade (P< 0.01), of which CALM3 was highly correlated with BM (P< 0.01). CALM3 mRNA was significantly down-regulated in BM (P< 0.05), and PRKCG mRNA was significantly down-regulated in both glioma and BM (P< 0.05).
Conclusion: The clinical invasive behavior between glioma and BM have significantly shortened the median survival time of patients. Down-regulation of CALM3 suggested a higher correlation with BM, meanwhile Down-regulation of PRKCG played a role in glioma and BM The detection of CALM3 and PRKCG may be helpful for invasive behavior and provide a reference for the targeted therapy in glioma and BM.
Keywords: Glioma; Brain metastasis; Bioinformatics; Neoplasm invasiveness
Introduction
Intracranial malignant tumor, characterized by its localization and local invasive growth, the disease exhibits high mortality and high disability rate. In general, intracranial malignant tumor can be divided into primary tumor and Brain Metastasis (BM).
Glioma accounts for nearly 30% of all primary brain tumors, and is the main cause of death of primary brain tumors [1]. The histological classification of gliomas is based on Bailey and Cushing’s embryonic theory and Kernohan’s anamorphology theory, which are astrocytoma, oligodendroglioma, oligoastrocytoma, ependymoma, and choroid plexus epithelioma. According to WHO’s central nervous system tumor classification in 2016, molecular typing is included in the diagnostic criteria for the first time, such as Isocitrate Dehydrogenase (IDH) wild/mutant, H3K27M mutant, 1p/19q codeletion, RELA fusion, WNT/SHH activation, TP 53 wild/mutant etc., performing individualized treatment and clinical prognosis analysis in patients [2]. However, the molecular typing and corresponding clinical significance of gliomas need to be further proved. The main reason is that clinical outcomes from current surgical outcomes, chemotherapy drugs, and radiotherapy, are not ideal [3]. The overall prognosis of patients with gliomas is still poor. The median survival time of glioblastoma is 1 year, and the 5-year mortality rate exceeds 95% [4]. With the deepening of research, most patients with gliomas have been confirmed to have polygenic mutations [5]. Therefore, it may provide guidance for improving the prognosis of glioma patients in studying the gene function network, finding a new gene mutation, using differentially expressed genes to add the molecular genetic diagnosis, and elaborating the potential regulatory mechanism.
Most neoplastic brain injuries are caused by cancers outside the central nervous system. Studies have shown that BM is the most common intracranial malignant tumor in adults. The incidence of peripheral BM is 10% -40%. The number of BM patients is about 10 times that of primary brain tumors. The median survival time of BM without treated is about 2 months, and can be extended to about 5 months after treatment [6]. The primary lesion of BM is lung cancer, followed by breast cancer, as well as malignant melanoma, gastrointestinal tumors, and kidney cancer [7]. Clinically, although BM patients have tried a variety of surgical-based treatment strategies, the results are not good [8]. By observing changed gene in BM tissues, and corresponding analysis with gliomas, it is possible to discover the underlying regulatory mechanism of BM, which has certain guiding significance for the clinical diagnosis and treatment [9].
At present, the genomic variation of gliomas is still a hot spot. The relational research can be divided into the following 7 groups [10]: telomerase reverse transcriptase (TERT, rs2736100), Epidermal growth factor receptor (EGFR), coiled-coil domain containing 26 mutant group (CCDC26, rs55705857), cyclin dependent kinase inhibitor 2B mutant group (CDKN2B, rs1412829), PH homologous domain B family member 1 mutation group (PHLDB1, rs498872), tumor protein p53 mutation group (TP53, rs78378222),and telomere extension Helicase regulator mutant group (RTEL1, rs6010620).The study of glioma genome variation not only analyzes the biological function and signaling pathway, but also guides clinical diagnosis and treatment plans. For example, the mutations of CCDC26 and PHLDB1 are associated with IDH variant gliomas, suggesting that patients have a lower relapse rate and longer survival time. The 1p/19q co-deletion is of great significance in the pathological diagnosis of oligodendroglioma, the evaluation of the efficacy of radiotherapy and chemotherapy, and the prediction of clinical prognosis, which is related to the variation of TERT. On the other hand, BM shows the characteristics of primary gene mutations, for example, nonsmall cell lung cancer (NSCLC) often takes EGFR mutation [11] and anaplastic lymphoma kinase gene (Anaplastic Lymphoma Kinase, ALK) Rearrangement. Breast cancer can be driven by human epidermal growth factor receptor 2 [12]. Melanoma often demonstrates BRAF V600E mutation.BM also illustrates intracranial malignant tumor characteristics in genes, such as TP53, NRAS, And KRAS mutations, DSC2, ST7, PIK3R1 and SMC mutations, etc. In short, genomic mutations of gliomas and BM provide ideas for new therapeutic targets.
Gene Sequencing, a high-throughput technology combining life science and microelectronics technology, has been widely studied and applied in various research fields of biology and medicine in recent years. It provides important theoretical and practical value for sequence analysis, gene expression, genome research, and hybridization signal intensity analysis with gene expression profiles. Accompanied large-scale, high-throughput information, a series of biological information in brain tumor research is integrated [13]. Zhu XP et al. [14] studied the expression characteristics of micro RNAs of different grades of gliomas through gene micro-matrix technology. Ondracek J et al. [15] discovered the full micro RNA expression profile of radio resistant glioblastoma cells. Bhawe KM et al. [16] formulated some principles of gene chip application in glioblastoma. Then, the expression profile of exons of gliomaswas found [17], and Protein- Protein Interactions (PPI) network construction and hierarchical cluster analysis were conducted [18]. It suggests the application of gene chip technology in the regulatory mechanism of gliomas.In addition, whole-gene high-throughput analysis of BM has also been reported [19]. However, there are few studies on the comparative analysis of differential genes between gliomas and BM.
This study collected samples and clinical data of glioma and BM, analyzing the clinical features of aggressive behavior, exploring the different genes between gliomas and BM by RNA sequencing (RNAseq), screening the invasive genes with bioinformatics principles, and finally verifying invasive genes by quantitative real-time polymerase chain reaction (qRT-PCR), which aimed at the molecular mechanism of intracranial malignant tumor progression.
Materials and Methods
Ethical Approval
The clinical data and tumor specimens pathologically diagnosed gliomas or metastases were collected from November 2017 to November 2019. The procedure was approved by the Ethics Committee of Guizhou Medical University and affiliated hospital (IRB-2019-152). All procedures performed in studies related to human participants were in accordance with the 1964 Declaration of Helsinki and its later amendments.
Clinical Data Recording and Grouping
52 patients were divided into low-grade glioma group (group A, n = 19), high-grade glioma group (group B, n = 18), BM group (group C, n = 15) .Gender, age, tumor location, tumor size, edema range, chief complaint, preoperative KPS, surgical approach, tumor blood supply, vascular invasion, presence or absence of cystic changes, degree of resection, tumor grade, and Ki 67 expression were recorded.
Specimen Collection and Processing
According to the operation plan, RNAwait (Solarbio, Beijing, China), cryotube, 4°C physiological saline, ice box, sterile instrument bag, sterile syringe, liquid nitrogen and liquid nitrogen were prepared 2h in advance. In the operating room environment, the tumor tissues (cancer tissues and adjacent tissues 3 cm away from the lesion center) were washed with normal saline, placed into cryotube in pieces, and put into a liquid nitrogen tank within 10 minutes. After 24 h, the tissues were transferred to a refrigerator at -80°C for future use.
RNA-seq
The RNA-seq process was supported by Shanghai Jinteda Gene Technology Co., Ltd (NO. G2016083036XM). Briefly, after conventional RNA extraction, Nanodrop 2000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA) determined the nucleic acid protein ratio A260/A280. The RNA concentration was measured on the Qubit 2.0 platform (Thermo Fisher Scientific, Waltham, Massachusetts, USA). 4200 TapeStation detected (Agilent Technologies, Santa Clara, California, USA) RNA RIN value. OligodT magnetic bead method was used to establish and amplify the mRNA library.
Bioinformatics Analysis
According to the previous report, the amplified sequences were analyzed using the next-generation sequencing comparison tools HISAT2 [20] and Stringtie [21]. Differentially expressed genes were obtained after correction using Fold-Change (FC). The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were used to enrich and annotate differentially expressed genes. After selection of invasion biological pathways involves genes, the reduced target invasion genes are confirmed by literature screening and human tissue expression.
qRT-PCR
cDNA was synthesized according to the instructions of the reverse transcription™ first strand cDNA synthesis Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Amplification was performed according to FastStart Universal SYBR Green Master (Roche, Basel, Switzerland). The reaction parameters set by the PCR instrument: pre-denaturation at 95°C for 2 min, denaturation at 95°C for 15 s, annealing for 20 s (Table 1) and extension at 60°C for 40 s. 40 cycles in total. after manual correction, the relative expression level were calculate by 2-ΔΔCt method. The sequence of the primers is shown in the (Table 1), synthesized by Beijing Solarbio company reference (http://asia.ensembl.org/).
gene
primer sequence
temperature
β -actin
sense5'- ATGTGCGACGAAGACGAGAC- 3'
54°C
antisense 5'-ACCAAGCAGGAGTACGACGA- 3
CALM3
sense5'- ATGGCTGACCAGCTGACTGA- 3'
51°C
antisense5'- TGGCCAGGTCAATTATGAAG- 3'
CAMK2A
sense5'- TGGCCACCATCACCTGCAC- 3'
50°C
antisense5'- TCGTCCACTTCCACAGATCT - 3'
CAMK2B
sense5'- ATGGCCACCACGGTGACCTG - 3'
53°C
antisense5'- AACGTGCACTTCCACTGCTC - 3'
PRKCG
sense5'- TGAGGCAGAAGGTGGTCCAC- 3'
54°C
antisense5'- CAGCGCTTCACCTACGTGAA - 3'
Table 1: Primer information of β -actin, CALM3, CAMK2A, CAMK2B, and PRKCG. Annealing temperature with β -actin was 54°C.Annealing temperature with CALM3 was 51°C.Annealing temperature with CAMK2A was 50°C. Annealing temperature with CAMK2B was 53°C.Annealing temperature with PRKCG was 54°C.
Follow-up
The follow-up started from the patient’s first diagnosis, by WeChat or telephone. The interval was 3 months. the deadline was 12 months, and the Overall Survival time (OS) was recorded.
Statistical Processing
IBM SPSS Statistics 20.0 was use for date processing. The measurement data adopts the mean ± standard deviation, and the count data is expressed by frequency (or percentage). Survival analysis draws Kaplan-Meier curve and performs log-rank test. Chisquare test (or Fisher’s exact test) was used for clinical data. Spearman correlation analysis was performed between invasion genes and tumor grade. Differences in the expression levels of the target genes in each group were analyzed by ANOVA. P<0.05 or P<0.01 means the difference is statistically significant.
Results
Poor Clinical Pathological Parameters Suggested Short OS
Univariate log-rank test analyzed the relationship between clinical characteristics and survival, and found that the median survival of patients who complained of non-headache was significantly longer than that of patients who complained of headache (11 months vs 7.5 months, P = 0.006).The median survival time of patients with tumor diameter <3 cm was significantly longer than that with tumor diameter ≥3 cm (11 months vs 5 months, P = 0.002). The median survival time of patients with tumor edema range <2 cm was significantly longer than that with tumor edema range ≥ 2 cm (12 months vs 3 months, P<0.01).The median survival time of patients with moderate tumor blood supply was significantly longer than that of tumor rich blood supply (11 months vs 5 months, P<0.01).
The median survival time of patients with tumors that did not invade blood vessels was significantly longer than those with tumors that invaded blood vessels (11 months vs 4 months, P<0.01).The median survival time of patients with complete tumor resection was significantly longer than that of patients with incomplete tumor resection (11 months vs 3 months, P<0.01).Patients with low-grade tumors had a significantly longer median survival than those with higher-grade tumors (12 months vs 8 months, P<0.01).The median survival time of patients with low expression of Ki 67 was significantly longer than that with high expression of Ki 67 (12 months vs 8 months, P<0.01).
Biological Enrichment of RNA-Seq Invasion Genes
After RNA-seq, the differentially expressed genes in the intersection of group A, B and C, a total of 1083 differentially expressed genes, of which 585 were up-regulated and 498 were down-regulated, were used for further screening. Through GO analysis of all upregulated differentially expressed genes, using standard enrichment methods, the selected biological pathways for invasion are: cell response to type I interferon, type I interferon signaling pathway, interferon-γ response, cell to interferon -γ response, Th1 and Th2 cell differentiation, NOD-like receptor signaling pathway, and Toll-like receptor signaling pathway. Comparatively, the biological pathways or signaling pathways for selected down-regulated differentially expressed genes are: glioma, Wnt signaling pathway, Rap1 signaling pathway. The above pathway further negative feedback to obtain the invasion genes involved.
Secondary Screening of Aggressive Differential Genes
According to the characteristics described in the above biological pathways or signaling pathways, the invasion genes can be screened into 5 group: Interferon-related genes, Th1 / Th2 balance genes, glioma genes, cell adhesion molecule genes, and pure signals pathway genes, as shown in Table 2. There are 66 candidate invasive genes screened by biological or signaling pathways, of which 48 are upregulated and 18 are down-regulated. Literatureswere searchedin PubMed database with the formula: (“Glioma” [Mesh] And “X gene” [gene]) or (“Neoplasm Metastasis” [Mesh] And “X gene” [gene]) (X represents the 66 genes). The selected literature standards are: 1) clinical specimens; 2) solid tumor samples; 3) there are clear experimental results about the X gene and invasiveness (tumor grade, lymphatic metastasis, prognosis, etc.) to illustrate the research progress of this gene. In order to explore the invasive genes that have not been studied, 32 genes were excluded, and a further 34 genes were obtained, of which 23 were up-regulated and 11 were downregulated. As shown in (Table 2).
gene
tumor type
n
conclusion
literature
CARD9
COAD
48
positively related to TNM
[22]
CASP1
glioma
6
up-regulation
[23]
CCL3L3
GBM
8
up-regulation
[24]
CCL8
PDAC
10
up-regulation
[25]
CD2
BRCA
819
well up-regulation
[26]
CD226
STAD
30
down-regulationof progression
[27]
CD274
glioma
21
up-regulation
[28]
CD4
NSCLC
102
positively related to TNM
[29]
CD8A
LUAD
49
up-regulation
[30]
CXCL10
CESC
60
down-regulationof invasion
[31]
CXCL9
SCC
51
up-regulationof progression
[32]
CYBB
GBP4
HLA-DOA
HLA-DQA1
HLA-DQA2
IFIT2
GBC
80
down-regulation
[33]
IFITM1
GBC
215
positively related to TNM
[34]
IFITM3
glioma
60
up-regulation
[35]
IFNA21
IL12RB1
IL4R
IL5
IRF5
BRCA
83
positively related tograde
[36]
IRF7
NPC
49
positively related tometastasis
[37]
LGALS9
glioma
1292
negatively related toOS
[38]
NFATC2
glioma
135
up-regulation
[39]
OAS2
COADREAD
130
positively related tometastasis
[40]
OAS3
OASL
PARP14
PARP9
PLCB2
RSAD2
SIGLEC1
PRAD
109
negatively related to invasion
[41]
SP100
GBM
19
down-regulation
[42]
SPN
GBM
20
up-regulation
[43]
STAT2
TBX21
BRCA
242
positively related toOS
[44]
TICAM2
TIGIT
TLR3
TLR7
CESC
126
up-regulation
[45]
TRIM21
TRIM22
TRPM2
UBD
glioma
165
positively related tograde
[46]
XAF1
OC
94
positively expression
[47]
ADCY1
AKT1
glioma
60
positively related tograde
[48]
CALM3
CAMK2A
CAMK2B
CNR1
glioma
16
down-regulation
[49]
DKK2
BRCA
98
down-regulation
[50]
GRIN2A
GRIN2B
P2RY1
PRKCG
RGS14
RSPO2
SCC
73
positively related to grade
[51]
THBS1
glioma
121
positively related to grade
[52]
WNT10B
PRAD
38
up-regulation
[53]
WNT11
OCC
47、127
positively related tometastasis
[54]
WNT16
WNT9A
COAD: Colon Adenocarcinoma; BRCA: Breast Invasive Carcinoma; STAD: Stomach Adenocarcinoma; LUAD: Lung Adenocarcinoma; GBC: Gallbladder Cancer; NPC: Nasopharyngeal Carcinoma; COADREAD: Colon Adenocarcinoma/Rectum Adenocarcinoma Esophageal Carcinoma; PRAD: Prostate Adenocarcinoma; OC: Ovarian Carcinoma; OCC: Ovarian and Cervical Carcinoma; GBM: Glioblastoma Multiforme; PDAC: Pancreatic Ductal Adenocarcinoma; NSCLC: Non-Small Cell Lung Cancer; CESC: Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma; SCC: Squamous Cell Carcinoma; OS: Overall Survival
Table 2: Statistics of aggressive differentially expressed genes. After biological pathways or signaling pathways screening, there are 66 candidate invasive genes, of which 48 are up-regulated and 18 are down-regulated. By PubMed screening, 34 genes were obtained, of which 23 were up-regulated and 11 were down-regulated. The clinical studies reported and the corresponding genes are shown in the table. The blank part indicates that it has not been reported yet.
CALM3, CAMK2A, CAMK2B, and PRKCG Were Responsible For the Invasion Genes
Searching the Gene Card database (https://www.GeneCards. org/), focusing on the distribution of 34 differentially expressed genes in human tissues and the sub cellular localization of the encoded protein, the down-regulated genes of CALM3, CAMK2A, CAMK2B, and PRKCG were verified. The four genes expression levels, mainly distributed in the brain and spinal cord, is more than twice that in other tissues or organs. Therefore, the targeted aggressive genes were theoretical confirmed.
CALM3, CAMK2A, CAMK2B, and PRKCG Shared Negative Correlation with Pathological Grade
Spearman correlation analysis of CALM3, CAMK2A, CAMK2B, PRKCG expression levels found that CALM3, CAMK2A, CAMK2B, PRKCG expression had a certain negative correlation with pathological grade. The lower the genes expressed, the higher the pathological grade was, and the stronger the aggressive behavior was. Among them, CALM3 expression level share no correlation in group B between group A, was moderately negatively correlated in group C between group B (P< 0.01, r = -0.716), and was highly negatively correlated in group C between group B (P< 0.01, r = -0.833). PRKCG expression levels were moderately negatively correlated in group B between group A (P< 0.01, r = -0.775), group C between group B (P< 0.01, r = -0.684), and in group C between B of analysis showed a high negative correlation (P< 0.01, r = -0.860).
Down-Regulation of CALM3 and PRKCG Was Responsible For Glioma and BM
Compared with group A, PRKCG mRNA expression level was significantly down-regulated (P<0.05). Compared with group A, the levels of CALM3 mRNA and PRKCG mRNA were significantly down-regulated (P<0.05). compared with group C and group B, the levels of CALM3 mRNA and PRKCG mRNA were significantly down-regulated (P<0.05) (Figure 1).
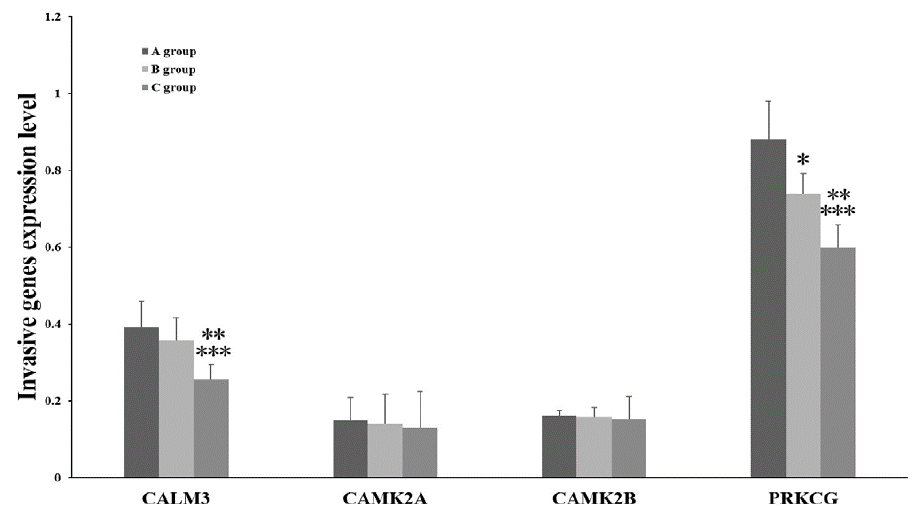
Figure 1: Compared with group A, PRKCG mRNA expression level was significantly down-regulated (P<0.05). Compared with group A, the levels of CALM3
mRNA and PRKCG mRNA were significantly down-regulated (P<0.05). Compared with group C and group B, the levels of CALM3 mRNA and PRKCG mRNA were
significantly down-regulated (P<0.05).
Discussion
Bioinformatics Shows More Practical Value In Intracranial Tumors
The basic process of bioinformatics in this study includes: 1) RNAGene seq; 2) preliminary screening of GO database and KEGG database; 3) further screening of PubMed database and GeneCard database. The key lies in the screening strategy or criteria [55]. The theory has always been that the brain is a relatively closed environment, which is not only anatomically closed, but also relatively closed in the blood system and immune system. Although in recent years, intracranial lymphatic vessels have been discovered, making it structurally possible for the immune escape theory, but compared with other organs outside the skull, physiological barriers, such as dura mater meninges, blood-brain barrier, and neuron gap junctions, naturally isolate many internal/external tumor communication channels. Intracranial tumors rarely spread to the extracranial, and extracranial tumors rarely spread to the intracranial [56,57]. In addition to the neuropathic factors, once the intracranial physiological barrier is destroyed, both intracranial tumors and extracranial tumors will show aggressive behavior, which is the particularity of intracranial tumors. Therefore, focusing on genes that are highly expressed in normal brain tissue is also a strategy for screening aggressive genes.
In recent years, the theoretical analysis of bioinformatics has exploded among intracranial tumors. Lingqi Zhou et al. [58] downloaded data from 79 samples (74 GBM and 5 normal tissues) in the Gene Expression Omnibus (GEO), and make theoretical prediction by using GO, KEGG, the database for annotation, visualization and integrated discovery (DAVID), PPI and UALCAN. 7 genes with high expression suggesting a good prognosis were identified. Xi X et al. [59] downloaded data from 701 samples (696 cases of gliomas and 5 normal tissues) of the cancer genome atlas (TCGA), using KEGG, PPI and weighted gene co -expression network analysis (WGCNA), and predicted four potential target genes for hsa-let-7b-5p treatment of glioma. Liu M et al. [60] downloaded data from 180 samples (26 astrocytomas, 50 oligodendrogliomas, 81 glioblastomas, and 23 normal tissues) in the GEO database. The theory predicts 4 genes with low expression suggesting a poor prognosis of glioma. Wei L et al. [61] downloaded data from 45 children’s tumor samples (35 diffuse gliomas, 10 normal tissues) in the GEO database. by prediction and verification in vitro with U251 tumor cell line, 4 genes and 1 miRNA were confirmed in the basic direction, and their high expression was related to the poor prognosis of the tumor. The specimens of this study are from clinical sources, mainly to explore aggressive genes, so the purpose and practicality may be better. Bioinformatics has always been based on computers. Whether it is an individual establishing a screening strategy or using a database on the Internet, there are varying degrees of reference value, but one thing to note is that it may be more effective combined with clinical analysis.
CALM3 and PRKCG Were More Related To the Invasiveness of Glioma and BM
In a spatial sense, low-grade gliomas were less aggressive than high-grade gliomas, and high-grade gliomas were less aggressive than metastatic tumors. The expression levels of CALM3 and PRKCG were higher in normal nerve tissues, but the expression levels in gliomas and BM were down-regulated in this study, which was negatively correlated with tumor aggressiveness. Further analysis showed that CALM3 mRNA was not significantly down-regulated in lowgrade gliomas and high-grade gliomas, but was significantly downregulated in BM, suggesting that CALM3 played an important role in the aggressive activity of BM. PRKCG mRNA was significantly down-regulatedin low-grade gliomas, high-grade gliomas, and BM, suggesting that PRKCG is associated with invasive behaviors in gliomas and BM.
CALM3, a calmodulin 3 gene, encoded a protein family member that binds calcium ions and functions as an enzymatic cofactor. The activity of this protein was important in the regulation of the cell cycle and cytokinesis, and its related pathways included ion channel transmission and protein metabolism [62]. CALM3 was associated with long Qt syndrome 1 and catecholaminergic polymorphism of ventricular tachycardia [63], and up-regulation of CALM3 in sleep deprived mice was also reported. This study found that CALM3 expression level was more down-regulated in BM, which may be related to calcium ion signaling pathway. PRKCG, protein kinase C γ gene,wasrelated to a serine/threonine-specific protein kinase that can be activated by calcium and second messengers [64]. This protein kinase was only expressed in the brain and spinal cord, and its localization is limited to neurons [65]. In this study, PRKCG has a certain correlation with gliomas and BM, which may play a role as a receptor or a second messenger in aggressive behavior. All above, It can be considered that this study was the first report that CALM3 and PRKCG were associated with gliomas and BM.
Conclusions
The clinical aggressive behavior of gliomas and BM shortens the median survival time of patients. Down-regulation of CALM3 was correlation with the aggressive behavior of BM, and down-regulation of PRKCG has a certain effect on the aggressive behavior of gliomas and BM. The detection of CALM3 and PRKCG may be helpful in evaluating the aggressive behavior of tumors and provide a reference for targeted therapy.
Acknowledgments
This work was supported in part by the Program for Changjiang Scholars and Innovative Research Team in University (IRT13058), National Natural Science Foundation of China(81560409), and the Program for Guizhou Province Science and Technology (LH-2016- 2905).The authors would like to thank the patients who have allowed her/his tumor tissues to be studied.
Funding Statement
This work was supported in part by the Program for Changjiang Scholars and Innovative Research Team in University (IRT13058), National Natural Science Foundation of China (81560409), and the Program for Guizhou Province Science and Technology (LH-2016- 2905).
References
- Hochberg FH, Atai NA, Gonda D, Hughes M, Mawejje B, et al. Glioma diagnostics and biomarkers: an ongoing challenge in the field of medicine and science. Expert review of molecular diagnostics. 2014; 14: 439-452.
- Louis DN, Perry A, Reifenberger G, Deimling A, Branger DF, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica. 2016; 131: 803-820.
- Chen R, Smith-Cohn M, Cohen AL, Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics: the journal of the American Society for Experimental Neuro Therapeutics. 2017; 14: 284-297.
- Zeng T, Cui D, Gao L. Glioma: an overview of current classifications, characteristics, molecular biology and target therapies. Frontiers in bioscience (Landmark edition). 2015; 20: 1104-1115.
- Oberndorfer S, Lindeck-Pozza E, Lahrmann H, Struhal W, Hitzenberger P, Grisold W. The end-of-life hospital setting in patients with glioblastoma. Journal of palliative medicine. 2008; 11: 26-30.
- Lin J, Jandial R, Nesbit A, Badie B, Chen M. Current and emerging treatments for brain metastases. Oncology (Williston Park, NY). 2015; 29: 250-257.
- Takei H, Rouah E, Ishida Y. Brain metastasis: clinical characteristics, pathological findings and molecular subtyping for therapeutic implications. Brain tumor pathology. 2016; 33: 1-12.
- Seoane J, De Mattos-Arruda L. Brain metastasis: new opportunities to tackle therapeutic resistance. Molecular oncology. 2014; 8: 1120-1131.
- Lowery FJ, Yu D. Brain metastasis: Unique challenges and open opportunities. Biochimica et biophysica acta Reviews on cancer. 2017; 1867: 49-57.
- van Breemen MS, Wilms EB, Vecht CJ. Epilepsy in patients with brain tumours: epidemiology, mechanisms, and management. The Lancet Neurology. 2007; 6: 421-430.
- Network CGAR. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014; 511: 543-550.
- Network. CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012; 490: 61-70.
- Du JY, Yang H, Tian DR, Wang QM, He L. Identification and functional analysis of differentially expressed genes related to obesity using DNA microarray. Genetics and molecular research: GMR. 2014; 13: 64-72.
- Zhu XP, Mou KJ, Xu QF, Tang JH, Huang GH, et al. Microarray analysis of the aberrant microRNA expression pattern in gliomas of different grades. Oncology reports. 2015; 34: 318-324.
- Ondracek J, Fadrus P, Sana J, Besse A, Loja T, et al. Global MicroRNA Expression Profiling Identifies Unique MicroRNA Pattern of Radioresistant Glioblastoma Cells. Anticancer research. 2017; 37: 1099-1104.
- Bhawe KM, Aghi MK. Microarray Analysis in Glioblastomas. Methods in molecular biology (Clifton, NJ). 2016; 1375: 195-206.
- Yu F, Fu WM. Identification of differential splicing genes in gliomas using exon expression profiling. Molecular medicine reports. 2015; 11: 843-850.
- He WQ, Gu JW, Li CY, Kuang YQ, Kong B, et al. The PPI network and clusters analysis in glioblastoma. European review for medical and pharmacological sciences. 2015; 19: 4784-4790.
- Saito N, Hatori T, Aoki K, Hayashi M, Hirata Y, et al. Dynamics of global gene expression changes during brain metastasis formation. Neuropathology: official journal of the Japanese Society of Neuropathology. 2009; 29: 389- 397.
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. 2015; 12: 357-360.
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. 2015; 33: 290-295.
- Yang M, Shao JH, Miao YJ, Cui W, Qi YF, et al. Tumor cell-activated CARD9 signaling contributes to metastasis-associated macrophage polarization. 2014; 21: 1290-1302.
- Jiang Z, Yao L, Ma H, Xu P, Li Z, et al. miRNA-214 Inhibits Cellular Proliferation and Migration in Glioma Cells Targeting Caspase 1 Involved in Pyroptosis. Oncology research. 2017; 25: 1009-1019.
- Zeiner PS, Preusse C, Golebiewska A, Zinke J, Iriondo A, et al. Distribution and prognostic impact of microglia/macrophage subpopulations in gliomas. Brain pathology (Zurich, Switzerland). 2019; 29: 513-529.
- Mou T, Xie F, Zhong P, Hua H, Lai L, et al. MiR-345-5p functions as a tumor suppressor in pancreatic cancer by directly targeting CCL8. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019; 111: 891-900.
- Han J, Choi YL, Kim H, Choi JY, Lee SK, et al. MMP11 and CD2 as novel prognostic factors in hormone receptor-negative, HER2-positive breast cancer. Breast cancer research and treatment. 2017; 164: 41-56.
- Han B, Mao FY, Zhao YL, Lv YP, Teng YS, et al. Altered NKp30, NKp46, NKG2D, and DNAM-1 Expression on Circulating NK Cells Is Associated with Tumor Progression in Human Gastric Cancer. 2018; 2018: 6248590.
- Wang Y, Wang L. miR-34a attenuates glioma cells progression and chemoresistance via targeting PD-L1. Biotechnology letters. 2017; 39: 1485- 1492.
- Zhang GQ, Han F, Fang XZ, Ma XM. CD4+, IL17 and Foxp3 expression in different pTNM stages of operable non-small cell lung cancer and effects on disease prognosis. Asian Pacific journal of cancer prevention : APJCP. 2012; 13: 3955-3960.
- Domagala-Kulawik J, Kwiecien I, Pankowski J, Pasieka-Lis M, Wolosz D, Zielinski M. Elevated Foxp3/CD8 Ratio in Lung Adenocarcinoma Metastatic Lymph Nodes Resected by Transcervical Extended Mediastinal Lymphadenectomy. BioMed research international. 2017; 2017: 5185034.
- Sato Y, Motoyama S, Nanjo H, Wakita A, Yoshino K, et al. CXCL10 Expression Status is Prognostic in Patients with Advanced Thoracic Esophageal Squamous Cell Carcinoma. Annals of surgical oncology. 2016; 23: 936-942.
- Li Z, Liu J, Li L, Shao S, Wu J, et al. Epithelial mesenchymal transition induced by the CXCL9/CXCR3 axis through AKT activation promotes invasion and metastasis in tongue squamous cell carcinoma. Oncology reports. 2018; 39: 1356-1368.
- Shen H, Zhan M, Zhang Y, Huang S, Xu S, et al. PLZF inhibits proliferation and metastasis of gallbladder cancer by regulating IFIT2. Cell death & disease. 2018; 9: 71.
- Li D, Yang Z, Liu Z, Zou Q, Yuan Y. DDR2 and IFITM1 Are Prognostic Markers in Gallbladder Squamous Cell/Adenosquamous Carcinomas and Adenocarcinomas. Pathology oncology research : POR. 2019; 25: 157-167.
- Zhao B, Wang H, Zong G, Li P. The role of IFITM3 in the growth and migration of human glioma cells. BMC neurology. 2013; 13: 210.
- Bi X, Hameed M, Mirani N, Pimenta EM, Anari J, Barnes BJ. Loss of interferon regulatory factor 5 (IRF5) expression in human ductal carcinoma correlates with disease stage and contributes to metastasis. Breast cancer research: BCR. 2011; 13: R111.
- Kondo S, Endo K, Wakisaka N, Aga M, KanoM, et al. Expression of interferon regulatory factor 7 correlates with the expression of Epstein-Barr Virus latent membrane protein 1 and cervical lymph node metastasis in nasopharyngeal cancer. Pathology international. 2017; 67: 461-466.
- Liang T, Wang X, Wang F, Feng E, You G. Galectin-9: A Predictive Biomarker Negatively Regulating Immune Response in Glioma Patients. World neurosurgery. 2019; 132: e455-e462.
- Tie X, Han S, Meng L, Wang Y, Wu A. NFAT1 is highly expressed in, and regulates the invasion of, glioblastoma multiforme cells. PloS one. 2013; 8: e66008.
- Kim JC, Ha YJ, Tak KH, Roh SA, Kwon YH, et al. Opposite functions of GSN and OAS2 on colorectal cancer metastasis, mediating perineural and lymphovascular invasion, respectively. PloS one. 2018; 13: e0202856.
- Stromvall K, Sundkvist K, Ljungberg B, Halin Bergstrom S, Bergh A. Reduced number of CD169(+) macrophages in pre-metastatic regional lymph nodes is associated with subsequent metastatic disease in an animal model and with poor outcome in prostate cancer patients. The Prostate. 2017; 77: 1468- 1477.
- Held-Feindt J, Hattermann K, Knerlich-Lukoschus F, Mehdorn HM, Mentlein R. SP100 reduces malignancy of human glioma cells. International journal of oncology. 2011; 38: 1023-1030.
- Buehler D, Waknitz M, Rehrauer W, Ranheim E, Selvaggi S. Small cell variant of malignant melanoma masquerading as lymphoma on fine-needle aspiration cytology: a case report. Diagnostic cytopathology. 2012; 40: 619- 623.
- Mori H, Kubo M, Kai M, Yamada M, Kurata K, et al. T-bet(+) lymphocytes infiltration as an independent better prognostic indicator for triple-negative breast cancer. Breast cancer research and treatment. 2019; 176: 569-577.
- Hasimu A, Ge L, Li QZ, Zhang RP, Guo X. Expressions of Toll-like receptors 3, 4, 7, and 9 in cervical lesions and their correlation with HPV16 infection in Uighur women. Chinese journal of cancer. 2011; 30: 344-350.
- Dai B, Zhang Y, Zhang P, Pan C, Xu C, et al. Upregulation of p-Smad2 contributes to FAT10-induced oncogenic activities in glioma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016; 37: 8621-8631.
- Wang Y, Liu P, Wang X, Mao H. Role of X-linked inhibitor of apoptosisassociated factor1 in vasculogenic mimicry in ovarian cancer. Molecular medicine reports. 2017; 16: 325-330.
- Chen L, Huang K, Han L, Shi Z, Zhang K, et al. beta-catenin/Tcf-4 complex transcriptionally regulates AKT1 in glioma. International journal of oncology. 2011; 39: 883-890.
- De Jesus ML, Hostalot C, Garibi JM, Salles J, Meana JJ, Callado LF. Opposite changes in cannabinoid CB1 and CB2 receptor expression in human gliomas. Neurochemistry international. 2010; 56: 829-833.
- Mu J, Hui T, Shao B, Li L, Du Z, et al. Dickkopf-related protein 2 induces G0/ G1 arrest and apoptosis through suppressing Wnt/beta-catenin signaling and is frequently methylated in breast cancer. Oncotarget. 2017; 8: 39443-39459.
- Zhang L, Song Y, Ling Z, Li Y, Ren X, et al. R-spondin 2-LGR4 system regulates growth, migration and invasion, epithelial-mesenchymal transition and stem-like properties of tongue squamous cell carcinoma via Wnt/betacatenin signaling. EBioMedicine. 2019; 44: 275-288.
- Daubon T, Leon C, Clarke K, Andrique L, Salabert L, et al. Deciphering the complex role of thrombospondin-1 in glioblastoma development. 2019; 10: 1146.
- Madueke I, hu W, Hu D, Swanson SM, Griend DV, et al. The role of WNT10B in normal prostate gland development and prostate cancer. The Prostate. 2019; 79.
- Azimian-Zavareh V, Hossein G, Ebrahimi M, Dehghani-Ghobadi Z. Wnt11 alters integrin and cadherin expression by ovarian cancer spheroids and inhibits tumorigenesis and metastasis. Experimental Cell Research. 2018; 369: 90-104.
- Silantyev AS, Falzone L, Libra M, Gurina OL, Kardashova KS, et al. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells. 2019; 8.
- Da Ros M, De Gregorio V, Iorio AL, Giunti L, Guidi M, et al. Glioblastoma Chemoresistance: The Double Play by Microenvironment and Blood-Brain Barrier. 2018; 19.
- Weidle UH, Niewohner J, Tiefenthaler G. The Blood-Brain Barrier Challenge for the Treatment of Brain Cancer, Secondary Brain Metastases, and Neurological Diseases. Cancer genomics & proteomics. 2015; 12: 167-177.
- Zhou L, Tang H, Wang F, Chen L, Ou S, et al. Bioinformatics analyses of significant genes, related pathways and candidate prognostic biomarkers in glioblastoma. Molecular medicine reports. 2018; 18: 4185-4196.
- Xi X, Chu Y, Liu N, Wang Q, Yin Z, et al. Joint bioinformatics analysis of underlying potential functions of hsa-let-7b-5p and core genes in human glioma. Journal of translational medicine. 2019; 17: 129.
- Liu M, Xu Z, Du Z, Wu B. The Identification of Key Genes and Pathways in Glioma by Bioinformatics Analysis. 2017; 2017: 1-9.
- Wei L, He F, Zhang W, Chen W, Yu B. Bioinformatics analysis of microarray data to reveal the pathogenesis of diffuse intrinsic pontine glioma. Biological research. 2018; 51: 26.
- Liu Y, Zheng X, Mueller GA, Zhang Y, London RE, et al. Crystal structure of calmodulin binding domain of orai1 in complex with Ca2+ calmodulin displays a unique binding mode. The Journal of biological chemistry. 2012; 287: 43030-43041.
- Gomez-Hurtado N, Boczek NJ, Kryshtal DO, Johnson CN, Sun J, et al. Novel CPVT-Associated Calmodulin Mutation in CALM3 (CALM3-A103V) Activates Arrhythmogenic Ca Waves and Sparks. Circulation Arrhythmia and electrophysiology. 2016; 9.
- Takahashi H, Adachi N, Shirafuji T, Danno S, Ueyama T, et al. Identification and characterization of PKCgamma, a kinase associated with SCA14, as an amyloidogenic protein. Human molecular genetics. 2015; 24: 525-539.
- Shirafuji T, Ueyama T. The Role of Cysteine String Protein alpha Phosphorylation at Serine 10 and 34 by Protein Kinase Cgamma for Presynaptic Maintenance. 2018; 38: 278-290.