Review Article
Austin J Clin Pathol. 2014;1(3): 1014.
Epigenetic Modifications and Carcinogenesis of Human Endometrial Cancer
Xianyong Ma*, Xiaobin Gao
Department of Pathology, Yale University School of Medicine, USA
*Corresponding author: Xianyong Ma, Department of Pathology, Yale University School of Medicine, New Haven, USA
Received: July 02, 2014; Accepted: Aug 01, 2014; Published: Aug 04, 2014
Introduction
The endometrial cancers (EC) or uterine cancers (UC), which arise from endometrium of uterus, are the seventh most common malignancies worldwide among females. According to the estimation from NCI, there will be around 52,630 new cases and 8,590 deaths from EC in USA in 2014 [1], therefore the exploration of the mechanisms for EC carcinogenesis and development for cost-effective treatment approaches are important and urgent. The significance of genetic alterations (changes DNA sequences) have been extensively explored in carcinogenesis of human endometrial cancer [2,3], but these studies do not provide a reasonable explanation of why the gene sequences are not changed in many endometrial cancer cases. Increasing evidence from recent research shows that the epigenetic regulation for gene expression is critical for endometrial carcinogenesis [4,5]. The epigenetic modifications do not change DNA sequences, but alter the side chain groups of DNA base or histone proteins, and then regulate gene expression to affect the biological function of cells. The epigenetic modifications can be structurally classified as following: 1. DNA methylations/demethylations; 2, Histone methylations/ demethylations; 3, Histone acetylations/deacetylations; 4, Histone phosphorelations/dephosphorelations; 5, other modifications such as deimination in DNA; β-N-acetylglucosamine, ADP ribosylation and Ubiquitylation/sumoylation in histones, also including histone tail clipping and histone proline isomerization. In addition, the concept of epigenetic regulation has been extended to microRNAs (miRNA) and LncRNA regulations, since these RNA molecules regulate the gene expression by partially match to target (complementary RNA strand) mRNA and then lead to inhibition and/or mRNA degradation. In this paper we review the impact of epigenetic modifications and related biological implications in carcinogenesis of human endometrial cancer, and also discuss possible treatment strategies based on the epigenetic alterations.
DNA Modifications in EC Carcinogenesis
In mammalian cells, DNA methylation/demethylation is one of the most popular epigenetic modifications and play fundamental role in regulation of gene expression.
The methylation status of promoter region determines if gene activation or inactivation, also control gene expression level. Abnormal DNA methylation patters (higher or lower than normal methylation level) have been associated with human tumors, as well as other neoplastic diseases [6].
Hypermethylation of tumor suppressor genes in EC carcinogenesis
The DNA methylation is catalyzed by DNA methyltransferases, which consist of three members of DNMT1, DNMT3A and DNMT3B. DNMT1 is the most abundant DNA methyltransferase among these enzymes. DNMT1 catalyzes the methylation of the 5'-cytosine in the CpG dinucleotide sequence, and plays an important role in maintaining the DNA methylation patterns during cell division [7]. The DNMT3A/3B catalyzes de novo methylation of DNA [8]. These three enzymes cooperatively catalyze the methylation reactions of CpG islands, which are often located in promoter regions of target genes [9]. Hypermethylation means the methylation exceeds physiological level of target DNAs (Figure 1), the hypermethylation of promoters leads to inactivate the expression of tumor suppressor genes and loss of corresponding proteins to repress carcinogenesis, thereby promoting carcinogenesis and enhancing the metastases of cancer cells. A number of tumor suppressor genes have been determined with frequent hypermethylation on promoter regions during endometrial carcinogenesis (See Table 1). The development of new assay methods for DNA methylation such as MLPA (methylation-specific multiplex ligation-dependent probe amplification)[10], Mass ARRAY analysis [11], MethylCap-Seq [12] supplies the possible tools to globally screen the methylations of tumor samples. MethylCap- Seq is based on the affinity purification of methylated CpG of DNA fragments by using tagged methylation binding protein MeCP2, and then sequencing the purified DNA fragments to understand the methylation status of DNA. Jones A et al. [13] carried out a global-scale hypermethylation assay to screen more than 27,000 CpG sites (from 64 endometrial cancers and 23 control samples), they found HAND2, which expresses in normal endometrium, is one of the most frequently hypermethylated and silenced genes in endometrial cancer cells. The global profiling for promoter methylation was also successfully used to find out hypermethylation statuses of tumor suppressor genes such as MLH1 [14], Tig, C/EBPα [15], PRs, ERs [16], microRNAs [17]. The assays for individual tumor suppressor genes have also been used to determine a number of genes that are repressed in endometrial cancer by hypermethylation of promoter regions, for example, the PTEN, a critical factor regulating PI3K-AKT pathway, has been detected with frequent mutations, deletion and promoter hypomethylation in EC [18], studies also showed that loss of PTEN expression due to promoter hypermethylation associated with MSI (Microsatellite instability) phenotype [19]. P16INK4a or CDKN2A, an inhibitor of cyclin-dependent kinases such as CDK4 and CDK6, plays as tumor suppressor in EC carcinogenesis, the promoter hypermethylation of P16INK4a gene has been reported in between 11% to 75% sporadic endometrial cancer [20-23]. Other tumor suppressor genes frequently detected with promoter hypermethylation in EC by similar procedures include RASSF1A (Ras associated domain gene family) (33%-85%) [24-25]; APC (Adenomatous polyposis coli) up to 46.6%) [26]; RUNX3 (86%) [27]; CDH13 (cadherin 13) (90%) [28]; E-cadherin (79.8%) [29]. In addition to these tumor suppressor genes, some potential tumor suppressor genes were also frequently detected: for example: 14-3-3s gene was hypermethylated in 40%-60% endometrial cancer and ovary cancer [30] (see Table 1 for detailed gene list).
DNA Methylation/Demethylation and Epigenetic Treatment of Cancers.
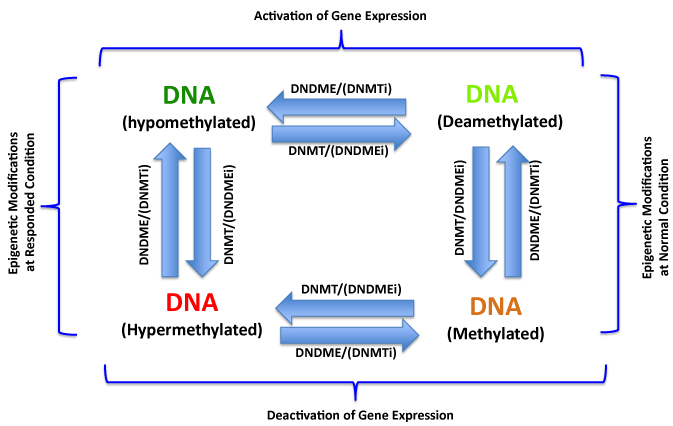
The DNA methylation/demethylation reactions catalyzed by DNA methyltransferase (DNMT) and demethylase (DNDME) respectively control DNA methylation status in cells. The hypermethylations of tumor suppressor gene promoters are very common epigenetic alterations and play critical role in endometrial carcinogenesis. The hypermethylation of DNA can be biochemically corrected by DNA methyltransferase inhibitors (DNMTi), therefore recovery the normal methylation status of tumor suppressor gene promoters and reactivate the expression of these genes to eventually overcome the malignant phenotype of cancer cells. Similarly, the DNA demethylase inhibitors (DNDMEi) inhibit the demethylation and inactivate these oncogenes in epigenetic treatment of EC and other cancers.
Figure 1:DNA Methylation/Demethylation and Epigenetic Treatment of Cancers.
The DNA methylation/demethylation reactions catalyzed by DNA methyltransferase (DNMT) and demethylase (DNDME) respectively control DNA methylation status in cells. The hypermethylations of tumor suppressor gene promoters are very common epigenetic alterations and play critical role in endometrial carcinogenesis. The hypermethylation of DNA can be biochemically corrected by DNA methyltransferase inhibitors (DNMTi), therefore recovery the normal methylation status of tumor suppressor gene promoters and reactivate the expression of these genes to eventually overcome the malignant phenotype of cancer cells. Similarly, the DNA demethylase inhibitors (DNDMEi) inhibit the demethylation and inactivate these oncogenes in epigenetic treatment of EC and other cancers.
Target Genes (Proteins)
Functions of Target Proteins
Hypermethyla-tion Locations
Regulation of Expression
Tumor Type
Reference
14-3-3 sigma
APC
AR
C/EBPa
CASP8
CDH1/E-cadherin
CDH13
CDKN2A/P16,
CHFR
CIDEA
COMT
EFEMP1
ERa
FHIT
GATA5
GSTP1
HAAO
HAND2
HOPX
MGMT
MiR129-2
MiR130a/b
MiR152
Mir196b
MiR200b
MiR203
miR222
MiR34b
MiR625
MLH1
MSH2
MT-1E
OX2R
P14-ARF
P73
Par-4
PEG3
PR
PR-B
PTEN
RAR-beta2
RASSF1A
RSK4
RXFP3
SFRP1
SPARC
Sprouty 2
TIG
TIMP3
TSLC1
VHL
WT1
ZNF154
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor **
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor
Tumor Suppressor P*
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
1st Exon
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
1stExon/1stIntron
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Promoter
Downregulation
Downregulation
Inactivation
Downregulation
Inactivation
Inactivation
Inactivation
Inactivation
Inactivation
Downregulation MSI
Inactivation
Inactivation
Inactivation
Inactivation
Inactivation
Inactivation
Downregulation MSI
Inactivation
Inactivation
Downregulation
Inactivation, MSI
Inactivation, MSI
Inactivation
Downregulation
Inactivation
Inactivation
Inactivation
Downregulation
Inactivation
Inactivation, MSI
Inactivation
Downregulation
Inactive
Inactivation
Downregulation
Downregulation, MSI
Inactivation
Inactivation
Inactivated
Inactivation, MSI
Downregulation
Inactivation
Inactivation
Downregulation MSI
Inactivation
Downregulation
Downregulation
Downregulation
Downregulation
Inactivation
Inactivation
Inactivation
Inactivation
EC, OC
EC
EC
EC
EC, OC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
ESC
EC
EC
EC
EC
EC
EC
EC
EC
EC, CC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC
EC, OC
EC, OC
EC
EC, OC
(30)
(21)
(67)
(15)
(68)
(24)
(28)
(20)
(69)
(73)
(70)
(71)
(16)
(29)
(72)
(24)
(73)
(13)
(74)
(75)
(72)
(77)
(77)
(17)
(77)
(77)
(77)
(78)
(77)
(77)
(79)
(80)
(81)
(82)
(83)
(84)
(85)
(86)
(87)
(19)
(88)
(24)
(89)
(73)
(86)
(90)
(89)
(15)
(91)
(92)
(68)
(94)
(68)
Table 1: Hypermethylation Genes in Endometrial Carcinogenesis.
Some members within same family can play bidirectional roles in endometrial carcinogenesis, for example: Homeobox (HOX) genes encode a group of homeodomain-containing transcription factors. Hox family genes have been identified as both tumor suppressor genes and oncogenes in carcinogenesis [31-33]. Some HOX members have been associated with endometrial cancer, Zhao et al. [34] reported that the HOXB13 was upregulated in endometrial cancer and possibly enhanced the invasiveness, which acts as an oncogenic protein in EC carcinogenesis, however, most of HOX family members act as tumor suppressors in EC carcinogenesis, for example, hypermethylation of Hoxa10 and Hoxa11 promoters are positively correlated with endometrial cancer types [35,36].
Hypomethylation of oncogenes in endometrial carcinogenesis
Similar with hypermethylation, the promoter hypomethylation (demethylation) is also a dynamic process presented in mammalian cells. In fact, during carcinogenesis, the DNA methylation pattern has paradoxical alteration: global DNA hypomethylation (demethylation) and local hypermethylation of certain genes. With almost all of the attention on the epigenetic modifications of DNA is focused on the promoter hypermethylations of tumor suppressor genes, very few publications have described the demethylation or hypomethylation in carcinogenesis for all cancers. Nonetheless, hypomethylation of oncogenes also play an important role in carcinogenesis as hypermethylation of tumor suppressor genes do.
In endometrial cancer, the hypomethylation of oncogenes is associated with early stage of carcinogenesis of endometrium through enhancing the ability of cell proliferation. Recently Erling et al. [37] reported CTCFL/BORIS gene (paralogue of CTCF-like factor, brother of the regulator of imprinted site) was hypomethylated on the promoter region and overexpression of this gene was significantly associated with endometrial tumorigenesis and poor survival of patients. Interestingly hypomethylations were also associated with the subtype of endometrial cancer, Hsu et al. [38] reported that bone morphogenetic protein members, BMP2, 3, 4, and 7, which were usually methylated in primary endometrial tumors with nonrecurrent type, but hypomethylated in primary endometrial tumors with the subsequent recurrent type, therefore the methylation patterns of BMP genes can be considered as the biomarkers of poor survival in endometrial cancer treatment. Some non-coding DNA sequences, such as the long interspersed element (LINE, the retrotransposons) [39], are hypomethylated in endometrial cancer, colorectal cancer and gastric cancer, and the hypomethylation of LINE-1 is also a putative biomarker for diagnosis of these cancers, however whether or not this gene directly acts as an oncogene during endometrial carcinogenesis remains unknown. The genes hypomethylated in endometrial cancer are listed in Table 2.
Target Genes (Proteins)
Functions of Target Proteins
Hypomethylation Locations
Regulation of Expression
Tumor Type
Reference
BMP 2,3,4,7
CASP8
CTCFL/BORIS
HOXB13
LICAM
LINE-1 **
MMP-2
PARP1
PAX2
S100A4
Oncogene
Oncogene-P*
Oncogene-P*
Oncogene-P*
Oncogene
Oncogene-P*
Oncogene-P*
Oncogene-P*
Oncogene
Oncogene
Promoter
Promoter
Promoter
Promoter
Promoter
Whole gene sequence
Promoter
Promoter
Promoter
1stIntron
Activation
Activation
Activation
Upregulation
Activation
Upregulation
Upregulation
Upregulation
Activation
Activation
EC
EC, OC
EC
EC
EC
EC
EC
EC
EC
EC
(38)
(68)
(37)
(34)
(95)
(39)
(96)
(97)
(98)
(99)
Table 2: Hypomethylation Genes in Endometrial Carcinogenesis.
Histone modifications in EC carcinogenesis
Histones are proteins that the DNA wraps itself to form chromatin. Tails of histone proteins are extensively modified post translationally in normal eukaryotic cells to maintain the functional structure of chromatin. Cancer cells frequently harbor aberrations in histones. Locus-specific alterations in histone modifications may have direct effects on expression of nearby genes. Moreover cancer cells also exhibit alterations in global level modifications, among these epigenetic modifications in histones, the acetylations/ deacetylations and methylations/ demethylations are the major histone modifications that have been reported to play a crucial role in carcinogenesis of EC.
Histone methylation and endometrial cancer
Methylations are one of the most frequent epigenetic modifications on core histones; the histone methylation is mediated by histone methyltransferase (HMT), which contain SET domain to catalyze the reaction of transferring methyl group from donor such as S-adenosyl methionine onto lysine or arginine residues of the H3 and H4 histones. The HMTs can be classified into two groups, group 1 (such as EZH2, Enhancer of Zeste Homolog 2) [40]) HMTs transfer methyl group to lysine residues of histone, group2 (such as PRMT, Protein Arginine Methyltransferase) HMTs transfer methyl group to arginine residues of H3 and H4 histones [41]. After the determination of biochemical function of chromatin repress complex (PRC2), whose core components include EZH2, EED and SUZ12. More and more researchers are focused on the study of EZH2 and PRC2, which transfers methyl group to lysine 27 and 9 residues of H3 [42]. Generally, the EZH2 acts as an oncoprotein during carcinogenesis, Yang et al. [43] found the EZH2 was upregulated in endometrial cancer resulting in hypermethylation of histone 3 lysine 27 of APC promoter, subsequently inactivated the expression of APC tumor suppressor. Zhou et al. [44] also reported that EZH2 was overexpressed in high-grade endometrial tumors. Therefore, the EZH2 was considered to play an oncogenic role in endometrial carcinogenesis by inhibition of tumor suppressor gene expression, even though the PRC2 complex is generally considered as a tumor suppressor complex. Due to the fact that PRC2 (with EZH2) can inactivate both oncogenes and tumor suppressor genes by methylation of histones, in the future, more mechanistic experiments need to be carried out to clarify the relationship of EZH2 oncogenic role and PRC2 tumor suppressor function.
Histone acetylation/deacetylation and endometrial cancer
Histone acetylation and deacetylation reactions, which are the important post-translational modifications involve in regulations of gene expression. Histone acetyltransferase (HAT) catalyzes histone acetylation by transfer acetyl group from the substrate acetyl-coenzyme A to histone, on the other hand, histone deacetylase (HDAC) catalyzes histone deacetylation by removing acetyl group from histone proteins via hydrolysis reaction (Figure 2). In the past years, a number of publications described the HAC activates the gene expression and HDAC acts the opposite, and both procedures play important roles in EC carcinogenesis [45,46].
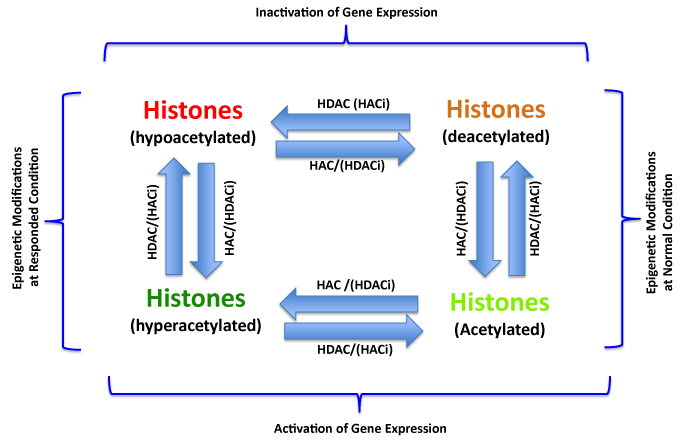
Histone Acetylation/Deacetylation and Epigenetic Treatment of Cancers.
The frequent aberrant histone modifications include hypoacetylations on tumor suppressor gene regions or hyperacetylations on oncogene regions in endometrial carcinogenesis. These procedures can be biochemically corrected by enzymatic inhibitors. The histone deacetylase inhibitor (HDACi) can inhibit the deacetylation and reactivation of tumor suppressor genes. Similarly, the acetyltransferase inhibitor (HACi) can inhibit the acetylation and inactivation of oncogenes, therefore, these inhibitors are considered to be as the drugs for EC or other cancer treatment.
Figure 2:Histone Acetylation/Deacetylation and Epigenetic Treatment of Cancers.
The frequent aberrant histone modifications include hypoacetylations on tumor suppressor gene regions or hyperacetylations on oncogene regions in endometrial carcinogenesis. These procedures can be biochemically corrected by enzymatic inhibitors. The histone deacetylase inhibitor (HDACi) can inhibit the deacetylation and reactivation of tumor suppressor genes. Similarly, the acetyltransferase inhibitor (HACi) can inhibit the acetylation and inactivation of oncogenes, therefore, these inhibitors are considered to be as the drugs for EC or other cancer treatment.
Histone deacetylation on tumor suppressor in endometrial carcinogenesis
Inactivation of tumor suppressor genes by deacetylation of histone is an important cause of carcinogenesis. A number of tumor suppressor genes have been identified to be associated with endometrial cancer (Table.3). In fact, a lot of evidence for inactivation of tumor suppressor genes by histone deacetylation was derived from those experiments that the deacetylation procedure is inhibited by HDAC inhibitors and resulted in reactivation of tumor suppressor genes [47-49]. Reactivated tumor suppressor gene in EC by HDAC inhibitor was also reported, Sarfstein [50] showed pTEN, P21 (WAF1) were upregulated in type I EC and IGF-IR and P21 were upregulated in type II EC when deacetylation was inhibited by HDAC inhibitor, these results indicated deacetylation of tumor suppressor genes involved in EC carcinogenesis. Interestingly, tumor suppressor gene p53 (in Ishikawa cells) and PTEN (in USPC-2 cells) were down regulated after treatment of these cells with HDAC inhibitor vorinostat, the mechanisms remain unknown, obviously, the HDAC inhibitors act with complicated patterns on the regulation of targets expression. In addition to the histone deacetylation on tumor suppressor in endometrial cancer, deacetylation of tumor suppressor genes are also involved in embryo invasion of endometrial stroma tissue, the TIPM- 1 and TIMP-3 were upregulated after treated endometrial stromal cells with HDAC inhibitor TSA, showed deacetylation of tumor suppressor genes exists not only in endometrial tumor cells but also in normal embryo invasion [51].
Target Genes/
(Proteins)
Functions of Target Proteins
Histone Modifications
Regulation of Expression
Tumor Type
Reference
APC
Bcl-2
C/EBPa
Caspase-9
CDKs
Cyclin A
Cyclin D1
E-Cadherin
Glycodelin
IGF-IR
MMP-2
MMP-9
P16
P21(WAF1)
P27
PR-B
pRb
pTEN,
TIG1
TIMP-1
TIMP-3
Tumor suppressor
Oncogene
Tumor suppressor
Tumor Suppressor*
Oncogene
Oncogene
Proto-oncogene
Tumor Suppressor
Tumor suppressor
Tumor Suppressor*
Oncogene
Oncogene
Tumor suppressor
Tumor suppressor
Tumor suppressor
Tumor suppressor
Tumor suppressor**
Tumor suppressor
Tumor suppressor *
Tumor suppressor
Tumor suppressor
Methylation
?
Deacetylation
Deacetylation
?
?
?
Deacetylation
Deacetylation
Deacetylation
?
?
Deacetylation
Deacetylation
Deacetylation
Decetylation
Deacetylation
Deacetylation
Deacetylation
Deacetylation
Deacetylation
Downregulation
Upregulation
Deactivation
Deaactivation
Upregulation
Activation
Upregulation
Downregulation
Deactivation
Downregulation
Activation
Activation
Deactivation
Deactivation
Deactivation
Deactivation
Upregulation
Up/Downregulation
Deactivation
Deactivation
Deactivation
EC
EC
EC
EC
EC
EC, OC
EC (type I, II)
EC, OC
EC
EC (type I, II)
EC
EC
EC, OC
EC
EC, OC
EC
EC
EC (type I, II)
EC
EC
EC
(43)
(66)
(13)
(93)
(49)
(100)
(50)
(100)
(101)
(50)
(51)
(51)
(100)
(50)
(100)
(102)
(49)
(50)
(15)
(51)
(51)
Table 3: Histone Modifications in Endometrial Carcinogenesis.
Histone acetylation on oncogene promoters in endometrial carcinogenesis
Acetylation catalyzes by a class of enzymes called histone acetyltransferases (HATs), HATs belong to a super family, which contains several subgroups as following: 1. Bromodomain-containing group: for instance: P300/CBP, TAFII250, GCN5, ATF2 and etc. [52]; 2. Chromodomain-containing group: Tip60, MOZ Sas3, MORF and etc. [53]; 3. Other family members, some nuclear cofactors such as SRC-1 [54], ACTR [55] and TIF-2 [56], these proteins also display HAT activity therefore is considered as members of HAT family. Similar with deacetylation catalyzed by deacetylase of tumor suppressor genes, theoretically the histone acetylation on oncogenes plays important role in carcinogenesis. A typical example derived from Kang et al. [57], they found HAT inhibitor "curcumin" which biochemically induces histone (H3/H4) hypoacetylation, downregulated the expression of oncogene PARP and Caspase-3, resulted in promotion of apoptosis, differentiation and effective neurogenesis. Unfortunately due to rare EC cases and few researchers are focused on the histone acetylation on oncogenes in EC, the direct evidence of relationship between acetylation on oncogenes and EC carcinogenesis is impossible at present, but some indirect data is still encouraging even there is confuse information, for example. Zhu et al. [58] found acetylation can inhibit the expression of proto-oncogenes such as IGBT3 in endometrial cancer cells (Ishikawa cells) via acetylating HOXA10 at K338 and K339. The acetylation status of core histones on promoter region of oncogenes in EC remains unknown. Undoubtedly, more work is needed to understand the pathological function of deacetylation for oncogenes, which activate through histone acetylation on promoter region in EC carcinogenesis. The acetylation/deacetylation of core histones are dynamic biochemical reactions in cells (Figure 2), the reversible and equilibrated reactions are maintained both by the regulations of network signaling pathways and feedback regulations triggered by microenvironment factors of cell, and also by the regulations of enzymatic kinetics (including substrate concentration and HAT activity). Based on gene expression patterns of cancer cells, any HDACi or HACi may act in a complicated pattern, in other words, one chemical compound modifies core histones at different epigenetic layers, depths and gene promoters depending on both the microenvironment regulation and structure/ conformational-specific interactions, therefore, causes multiple and even opposite results at exteriorly.
The histone acetylation and promoter DNA hypomethylation synergistically activate the oncogene expression in carcinogenesis of endometrial cancer and other cancer types, experiments showed only acetylated histones bind to unmethylated MLH1 promoters, that indicated both of promoter activation by hypomethylation and histone activation by acetylation are required for gene expression [13,59,60].
Epigenetic modification and treatment strategy for endometrial cancer
Treatment of endometrial cancer at epigenetic level by small chemical compounds can partially or even fully restore normal expression level of functional genes including upregulation of inactivated tumor suppressor genes and deactivation of activated oncogenes. At present, inhibition of HDAC activity is one of the most common method for epigenetic treatment of cancers including endometrial cancer, and a number of HDAC inhibitors are developed and being used on clinic trials, some drugs are already into clinic phase III such as TSA (Trichostatin A) [61], which inhibits almost all of the HDAC members but HDAC8, the biological function of this epigenetic drug includes: 1, induce apoptosis of cancer cells; 2, induce cancer cell differentiation. Panobinostat is a novel HDAC inhibitor with a broad-spectrum HDAC inhibition activity developed by Novartis Company and has been used to against many type of cancers [62], this drug has been shown to inhibit proliferation and induce apoptosis of EC cells. Tacedinaline (CI994) is also a HDAC inhibitor newly developed by Pfizer Company that has been used in phase III clinical trial [63], and the biological function for this drug included mediation of G1 cell cycle arrest, inhibition of proliferation and induction of apoptosis of cancer cells. At present more than 30 compounds are being used for preclinical or early stage clinical trials for the inhibition of HDAC activity such as Dacinostat, Sodium phenylbutyrate, CUDC-907, Tubastatin, Tubacin, Givinostat.
Since silence of tumor suppressor genes by promoter hypermethylation also plays an important role in the development and progression of EC, therefore another valuable target for epigenetic therapy of endometrial cancer is to directly inhibit DNA methyltransferase activity. At present, the most popular DNMT inhibitors (DNMTi) both for experimental and clinical purpose are cytidine analogues such as 5-azacytidine, 5-aza-2'-deoxycytidine (decitabine) and pyrimidin-2-one ribonucleoside (zebularine). Experiment showed that DNMTi (decitabine) induces apoptosis, growth inhibition and G2 arrest in human endometrial cancer cells [64]. The combination of DNMTi and HDACi as a new treatment strategy showed better efficacy to cancer patients compared with separated administration. Xu et al [65] recently reported the presence of strong synergistic effect between DMNTi (ADC) and HDACi (TSA), TSA appeared to be a more potent apoptosis inducer, but have smaller effect on cell cycle, in the reverse effect; ADC exhibited strong regulation on cell cycle, but had smaller effect on apoptosis. Yi et al. [66] showed combination of DNMTi (ADC) and HDACi (VPA, valproic acid) treatment inhibited tumor growth of endometrial cell lines (HEC1B), and upregulated CDH1 and downregulated Bcl-2 expression levels. Doubtlessly, treatment of endometrial cancer with administration of both inhibitors for DNMTs and for HDACs should be one of the major methods for epigenetic therapy of endometrial cancer in the future.
Summary
It becomes clear that the initiation, progression and metastasis of endometrial cancer are controlled both by genetic and epigenetic events. Genetic alterations associated with EC carcinogenesis involve several critical genetic events such as high frequent mutations of PTEN, K-RAS, P53 etc., and these genetic changes result in interfering corresponding signaling pathways (for examples: PI3K/AKT/mTOR; WNT/β-catenin, MAPK/ERK and etc.), and thereafter the cells obtain the transforming capabilities to form the endometrial cancer phenotype. At the layer of modifications for DNA and core histones, epigenetic alterations also play an important role in EC carcinogenesis (Figure 3), for most EC cases, the genetic and epigenetic alterations are both existed and may have synergistic effect, for example: PTEN acts as a key mediator of signaling pathway involved in endometrial cancer due to frequent mutations, interestingly, this gene has also been detected with histone deacetylation on promoter region for these endometrial cancer cases. Other genes such as P16 or pRB were also frequently detected with genetic alterations or epigenetic alterations. These alterations at genetic and epigenetic levels show a potential, encouraging possibility for endometrial cancer treatment, which means by targeting both levels, the malignant phenotype can be reverted to normal state through adjusting the expression of target genes at epigenetic layer.
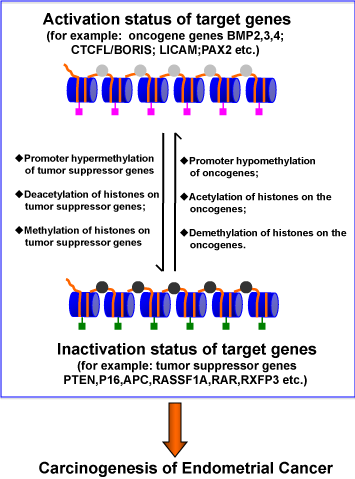
The Epigenetic Modifications and Endometrial Carcinogenesis.
The chromatin consists of the nucleosome, which comprises a sequence of DNA wrapped around core histones. The cis elements existed in DNA sequences such as promoters are abnormally modified including promoter hypermethylation of tumor suppressor genes, promoter hypomethylation of oncogenes. On the other hand, the aberrant core histone modifications such as deacetylation or site- specific methylations of histones on tumor suppressor genes, demethylation or acetylations of histone on oncogenes are co-operationally interactions with DNA alterations to interfere the gene expression and promote the carcinogenesis of endometrial cancer.
Figure 3:The Epigenetic Modifications and Endometrial Carcinogenesis.
The chromatin consists of the nucleosome, which comprises a sequence of DNA wrapped around core histones. The cis elements existed in DNA sequences such as promoters are abnormally modified including promoter hypermethylation of tumor suppressor genes, promoter hypomethylation of oncogenes. On the other hand, the aberrant core histone modifications such as deacetylation or site- specific methylations of histones on tumor suppressor genes, demethylation or acetylations of histone on oncogenes are co-operationally interactions with DNA alterations to interfere the gene expression and promote the carcinogenesis of endometrial cancer.
The methylation/demethylation of DNA and acetylation/ deacetylation of histones are the most common and important epigenetic modifications in normal and cancer cells. Critical issues for exploring epigenetic alterations will need to understand the following aspects: 1, understand how global hypomethylations and local hypermethylations (located in promoter region of tumor suppressor genes) take place in carcinogenesis; 2, how to regulate the reversible reactions for these epigenetic modifications in normal and cancer cells; 3, the most challenging work is site- or position-specific modifications for the DNAs or histones in chromatin, since epigenetic drugs (HDACi, DMNTi) may randomly reactivate tumor suppressor genes and also activate proto-oncogenes at same time; 4, explore the relationships between daily nutrients containing methyl or acetyl groups which are critical substrates for epigenetic modifications and may involve in carcinogenesis or anti-carcinogenesis.
Acknowledgment
We are very grateful to Dr. Jeffrey Sklar for reading the manuscript and his critical comments.
References
- American Cancer Society: Cancer Facts and Figures 2014. Atlanta, Ga: American Cancer Society. 2014.
- Yeramian A, Moreno-Bueno G, Dolcet X, Catasus L, Abal M, Colas E, et al. Endometrial carcinoma: molecular alterations involved in tumor development and progression. See comment in PubMed Commons below Oncogene. 2013; 32: 403-413.
- Llobet D, Pallares J, Yeramian A, Santacana M, Eritja N, Velasco A, et al. Molecular pathology of endometrial carcinoma: practical aspects from the diagnostic and therapeutic viewpoints. See comment in PubMed Commons below J Clin Pathol. 2009; 62: 777-785.
- Sakuragi N. Recent advances in research on epigenetic alterations and clinical significance of para-aortic lymphadenectomy in endometrial cancer: an introduction. International journal of clinical oncology. 2013; 18: 183-185.
- Arafa M, Somja J, Dehan P, Kridelka F, Goffin F, Boniver J, et al. Current concepts in the pathology and epigenetics of endometrial carcinoma. See comment in PubMed Commons below Pathology. 2010; 42: 613-617.
- Tao MH, Freudenheim JL. DNA methylation in endometrial cancer. See comment in PubMed Commons below Epigenetics. 2010; 5: 491-498.
- Szyf M, Bozovic V, Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. See comment in PubMed Commons below J Biol Chem. 1991; 266: 10027-10030.
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. See comment in PubMed Commons below Cell. 1999; 99: 247-257.
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. See comment in PubMed Commons below Nat Rev Genet. 2012; 13: 484-492.
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, et al. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. See comment in PubMed Commons below Nucleic Acids Res. 2005; 33: e128.
- Jurinke C, Denissenko MF. A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mutation research. 2005; 573: 83-95.
- Bock C, Tomazou EM, Brinkman AB, Müller F, Simmer F, Gu H, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. See comment in PubMed Commons below Nat Biotechnol. 2010; 28: 1106-1114.
- Jones A, Teschendorff AE, Li Q, Hayward JD, Kannan A, Mould T, et al. Role of DNA methylation and epigenetic silencing of HAND2 in endometrial cancer development. See comment in PubMed Commons below PLoS Med. 2013; 10: e1001551.
- Xiong Y, Dowdy SC. hMLH1 promoter methylation and silencing in primary endometrial cancers are associated with specific alterations in MBDs occupancy and histone modifications. Gynecologic oncology. 2006; 103: 321-328.
- Takai N, Kawamata N, Walsh CS, Gery S, Desmond JC, Whittaker S, et al. Discovery of epigenetically masked tumor suppressor genes in endometrial cancer. See comment in PubMed Commons below Mol Cancer Res. 2005; 3: 261-269.
- Ghabreau L, Roux JP, Niveleau A, Fontaniére B, Mahe C, Mokni M, et al. Correlation between the DNA global methylation status and progesterone receptor expression in normal endometrium, endometrioid adenocarcinoma and precursors. Virchows Archiv: an international journal of pathology. 2004; 445: 129-134.
- Abe W, Nasu K, Nakada C, Kawano Y, Moriyama M, Narahara H, et al. miR-196b targets c-myc and Bcl-2 expression, inhibits proliferation and induces apoptosis in endometriotic stromal cells. See comment in PubMed Commons below Hum Reprod. 2013; 28: 750-761.
- Dellas A, Jundt G. Combined PTEN and p27kip1 protein expression patterns are associated with obesity and prognosis in endometrial carcinomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 2009; 15: 2456-2462.
- Salvesen HB, MacDonald N, Ryan A, Jacobs IJ, Lynch ED, Akslen LA, et al. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. International journal of cancer Journal international du cancer. 2001; 91: 22-26.
- Guida M, Sanguedolce F, Bufo P, Di Spiezio Sardo A, Bifulco G, Nappi C, et al. Aberrant DNA hypermethylation of hMLH-1 and CDKN2A/p16 genes in benign, premalignant and malignant endometrial lesions. See comment in PubMed Commons below Eur J Gynaecol Oncol. 2009; 30: 267-270.
- Wong YF, Chung TK, Cheung TH, Nobori T, Yu AL, Yu J, et al. Methylation of p16INK4A in primary gynecologic malignancy. See comment in PubMed Commons below Cancer Lett. 1999; 136: 231-235.
- Furlan D, Carnevali I, Marcomini B, Cerutti R, Dainese E, Capella C, et al. The high frequency of de novo promoter methylation in synchronous primary endometrial and ovarian carcinomas. See comment in PubMed Commons below Clin Cancer Res. 2006; 12: 3329-3336.
- Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. See comment in PubMed Commons below BMC Cancer. 2006; 6: 212.
- Fiolka R, Zubor P, Janusicova V, Visnovsky J, Mendelova A, Kajo K, et al. Promoter hypermethylation of the tumor-suppressor genes RASSF1A, GSTP1 and CDH1 in endometrial cancer. See comment in PubMed Commons below Oncol Rep. 2013; 30: 2878-2886.
- Kang S, Kim JW, Kang GH, Lee S, Park NH, Song YS, et al. Comparison of DNA hypermethylation patterns in different types of uterine cancer: cervical squamous cell carcinoma, cervical adenocarcinoma and endometrial adenocarcinoma. International journal of cancer Journal international du cancer. 2006; 118: 2168-2171.
- Moreno-Bueno G, Hardisson D, Sánchez C, Sarrió D, Cassia R, García-Rostán G, et al. Abnormalities of the APC/beta-catenin pathway in endometrial cancer. See comment in PubMed Commons below Oncogene. 2002; 21: 7981-7990.
- Yoshizaki T, Enomoto T, Fujita M, Ueda Y, Miyatake T, Fujiwara K, et al. Frequent inactivation of RUNX3 in endometrial carcinoma. See comment in PubMed Commons below Gynecol Oncol. 2008; 110: 439-444.
- Seeber LM, Zweemer RP, Marchionni L, Massuger LF, Smit VT, van Baal WM, et al. Methylation profiles of endometrioid and serous endometrial cancers. See comment in PubMed Commons below Endocr Relat Cancer. 2010; 17: 663-673.
- Leal Rojas P, Anabalón Rodríguez L, García Muñoz P, Tapia Escalona O, Guzmán González P, Araya Orostica JC, et al. [Promoter hypermethylation gene patterns in gynecological tumors]. See comment in PubMed Commons below Med Clin (Barc). 2009; 132: 371-376.
- Mhawech P, Benz A, Cerato C, Greloz V, Assaly M, Desmond JC, et al. Downregulation of 14-3-3sigma in ovary, prostate and endometrial carcinomas is associated with CpG island methylation. See comment in PubMed Commons below Mod Pathol. 2005; 18: 340-348.
- Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. See comment in PubMed Commons below Nat Rev Cancer. 2010; 10: 361-371.
- Velu CS, Chaubey A, Phelan JD, Horman SR, Wunderlich M, Guzman ML, et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. See comment in PubMed Commons below J Clin Invest. 2014; 124: 222-236.
- Delval S, Taminiau A, Lamy J, Lallemand C, Gilles C, Noël A, et al. The Pbx interaction motif of Hoxa1 is essential for its oncogenic activity. See comment in PubMed Commons below PLoS One. 2011; 6: e25247.
- Zhao Y, Yamashita T, Ishikawa M. Regulation of tumor invasion by HOXB13 gene overexpressed in human endometrial cancer. See comment in PubMed Commons below Oncol Rep. 2005; 13: 721-726.
- Yoshida H, Broaddus R, Cheng W, Xie S, Naora H. Deregulation of the HOXA10 homeobox gene in endometrial carcinoma: role in epithelial-mesenchymal transition. See comment in PubMed Commons below Cancer Res. 2006; 66: 889-897.
- Whitcomb BP, Mutch DG, Herzog TJ, Rader JS, Gibb RK, Goodfellow PJ, et al. Frequent HOXA11 and THBS2 promoter methylation, and a methylator phenotype in endometrial adenocarcinoma. See comment in PubMed Commons below Clin Cancer Res. 2003; 9: 2277-2287.
- Hoivik EA, Kusonmano K, Halle MK, Berg A, Wik E, Werner HM, et al. Hypomethylation of the CTCFL/BORIS promoter and aberrant expression during endometrial cancer progression suggests a role as an Epi-driver gene. Oncotarget. 2014; 5: 1052-1061.
- Hsu YT, Gu F, Huang YW, Liu J, Ruan J, Huang RL, et al. Promoter hypomethylation of EpCAM-regulated bone morphogenetic protein gene family in recurrent endometrial cancer. See comment in PubMed Commons below Clin Cancer Res. 2013; 19: 6272-6285.
- Pavicic W, Joensuu EI, Nieminen T, Peltomäki P. LINE-1 hypomethylation in familial and sporadic cancer. See comment in PubMed Commons below J Mol Med (Berl). 2012; 90: 827-835.
- Trievel RC, Flynn EM, Houtz RL, Hurley JH. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. See comment in PubMed Commons below Nat Struct Biol. 2003; 10: 545-552.
- Thomas D, Lakowski TM. Forster resonance energy transfer measurements of cofactor-dependent effects on protein arginine N-methyltransferase homodimerization. Protein Sci. 2010; 19: 2141-2151.
- Breuer RH, Snijders PJ, Smit EF, Sutedja TG, Sewalt RG, Otte AP, et al. Increased expression of the EZH2 polycomb group gene in BMI-1-positive neoplastic cells during bronchial carcinogenesis. Neoplasia. 2004; 6: 736-743.
- Yang Y, Zhou L, Lu L, Wang L, Li X, Jiang P, et al. A novel miR-193a-5p-YY1-APC regulatory axis in human endometrioid endometrial adenocarcinoma. See comment in PubMed Commons below Oncogene. 2013; 32: 3432-3442.
- Zhou J, Roh JW, Bandyopadhyay S, Chen Z, Munkarah AR, Hussein Y, et al. Overexpression of enhancer of zeste homolog 2 (EZH2) and focal adhesion kinase (FAK) in high grade endometrial carcinoma. See comment in PubMed Commons below Gynecol Oncol. 2013; 128: 344-348.
- Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. See comment in PubMed Commons below Mol Oncol. 2007; 1: 19-25.
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. See comment in PubMed Commons below Cell. 1999; 98: 675-686.
- Feng D, Cao Z, Li C, Zhang L, Zhou Y, Ma J, et al. Combination of valproic acid and ATRA restores RARβ2 expression and induces differentiation in cervical cancer through the PI3K/Akt pathway. See comment in PubMed Commons below Curr Mol Med. 2012; 12: 342-354.
- Tran HT, Kim HN, Lee IK, Nguyen-Pham TN, Ahn JS, Kim YK, et al. Improved therapeutic effect against leukemia by a combination of the histone methyltransferase inhibitor chaetocin and the histone deacetylase inhibitor trichostatin A. Journal of Korean medical science. 2013; 28: 237-246.
- Ahn MY, Jung JH, Na YJ, Kim HS. A natural histone deacetylase inhibitor, Psammaplin A, induces cell cycle arrest and apoptosis in human endometrial cancer cells. See comment in PubMed Commons below Gynecol Oncol. 2008; 108: 27-33.
- Sarfstein R, Bruchim I, Fishman A, Werner H. The mechanism of action of the histone deacetylase inhibitor vorinostat involves interaction with the insulin-like growth factor signaling pathway. See comment in PubMed Commons below PLoS One. 2011; 6: e24468.
- Estella C, Herrer I, Atkinson SP, Quiñonero A, Martínez S, Pellicer A, et al. Inhibition of histone deacetylase activity in human endometrial stromal cells promotes extracellular matrix remodelling and limits embryo invasion. PloS one. 2012; 7: e30508.
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic acids research. 2002; 30: 5036-5055.
- Smith AT, Tucker-Samaras SD, Fairlamb AH, Sullivan WJ Jr. MYST family histone acetyltransferases in the protozoan parasite Toxoplasma gondii. See comment in PubMed Commons below Eukaryot Cell. 2005; 4: 2057-2065.
- Spencer TE, Jenster G, Burcin MM, Allis CD, Zhou J, Mizzen CA, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. See comment in PubMed Commons below Nature. 1997; 389: 194-198.
- Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. See comment in PubMed Commons below Nature. 1998; 394: 498-502.
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. See comment in PubMed Commons below Microbiol Mol Biol Rev. 2000; 64: 435-459.
- Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. See comment in PubMed Commons below Stem Cells Dev. 2006; 15: 165-174.
- Zhu LH, Sun LH, Hu YL, Jiang Y, Liu HY, Shen XY, et al. PCAF impairs endometrial receptivity and embryo implantation by down-regulating β3-integrin expression via HOXA10 acetylation. See comment in PubMed Commons below J Clin Endocrinol Metab. 2013; 98: 4417-4428.
- Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. See comment in PubMed Commons below Mol Cancer Res. 2006; 4: 339-349.
- Brodie SA, Li G, El-Kommos A, Kang H, Ramalingam SS, Behera M, et al. Class I HDACs are mediators of smoke carcinogen-induced stabilization of DNMT1 and serve as promising targets for chemoprevention of lung cancer. See comment in PubMed Commons below Cancer Prev Res (Phila). 2014; 7: 351-361.
- Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC, et al. Clinical development of histone deacetylase inhibitors as anticancer agents. See comment in PubMed Commons below Annu Rev Pharmacol Toxicol. 2005; 45: 495-528.
- Prince HM, Bishton MJ, Johnstone RW. Panobinostat (LBH589): a potent pan-deacetylase inhibitor with promising activity against hematologic and solid tumors. See comment in PubMed Commons below Future Oncol. 2009; 5: 601-612.
- Beckers T, Burkhardt C, Wieland H, Gimmnich P, Ciossek T, Maier T, et al. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. International journal of cancer Journal international du cancer. 2007; 121: 1138-1148.
- Cui M, Wen Z, Chen J, Yang Z, Zhang H. 5-Aza-2'-deoxycytidine is a potent inhibitor of DNA methyltransferase 3B and induces apoptosis in human endometrial cancer cell lines with the up-regulation of hMLH1. See comment in PubMed Commons below Med Oncol. 2010; 27: 278-285.
- Xu S, Ren J, Chen HB, Wang Y, Liu Q, Zhang R. Cytostatic and apoptotic effects of DNMT and HDAC inhibitors in endometrial cancer cells. See comment in PubMed Commons below Curr Pharm Des. 2014; 20: 1881-1887.
- Yi TZ, Li J, Han X, Guo J, Qu Q, Guo L, et al. DNMT inhibitors and HDAC inhibitors regulate E-cadherin and Bcl-2 expression in endometrial carcinoma in vitro and in vivo. See comment in PubMed Commons below Chemotherapy. 2012; 58: 19-29.
- Sasaki M, Oh BR, Dharia A, Fujimoto S, Dahiya R. Inactivation of the human androgen receptor gene is associated with CpG hypermethylation in uterine endometrial cancer. See comment in PubMed Commons below Mol Carcinog. 2000; 29: 59-66.
- Sánchez-Vega F, Gotea V, Petrykowska HM, Margolin G, Krivak TC, DeLoia JA, et al. Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics: official journal of the DNA Methylation Society. 2013; 8: 1355-1372.
- Wang X, Yang Y, Xu C, Xiao L, Shen H, Zhang X, et al. CHFR suppression by hypermethylation sensitizes endometrial cancer cells to paclitaxel. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2011; 21: 996-1003.
- Sasaki M, Kaneuchi M, Sakuragi N, Dahiya R. Multiple promoters of catechol-O-methyltransferase gene are selectively inactivated by CpG hypermethylation in endometrial cancer. See comment in PubMed Commons below Cancer Res. 2003; 63: 3101-3106.
- Yang T, Qiu H, Bao W, Li B, Lu C, Du G, et al. Epigenetic inactivation of EFEMP1 is associated with tumor suppressive function in endometrial carcinoma. See comment in PubMed Commons below PLoS One. 2013; 8: e67458.
- Dvorakova E, Chmelarova M. Methylation analysis of tumor suppressor genes in endometroid carcinoma of endometrium using MS-MLPA. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia. 2013; 157: 298-303.
- Huang YW, Luo J, Weng YI, Mutch DG, Goodfellow PJ, Miller DS, et al. Promoter hypermethylation of CIDEA, HAAO and RXFP3 associated with microsatellite instability in endometrial carcinomas. See comment in PubMed Commons below Gynecol Oncol. 2010; 117: 239-247.
- Yamaguchi S, Asanoma K. Homeobox gene HOPX is epigenetically silenced in human uterine endometrial cancer and suppresses estrogen-stimulated proliferation of cancer cells by inhibiting serum response factor. International journal of cancer Journal international du cancer. 2009; 124: 2577-2588.
- Nagy E, Gajjar KB, Patel II, Taylor S, Martin-Hirsch PL, Stringfellow HF, et al. MGMT promoter hypermethylation and K-RAS, PTEN and TP53 mutations in tamoxifen-exposed and non-exposed endometrial cancer cases. See comment in PubMed Commons below Br J Cancer. 2014; 110: 2874-2880.
- Huang YW, Kuo CT, Chen JH, Goodfellow PJ, Huang TH, Rader JS, et al. Hypermethylation of miR-203 in endometrial carcinomas. See comment in PubMed Commons below Gynecol Oncol. 2014; 133: 340-345.
- Li BL, Lu W, Lu C, Qu JJ, Yang TT, Yan Q, et al. CpG island hypermethylation-associated silencing of microRNAs promotes human endometrial cancer. See comment in PubMed Commons below Cancer Cell Int. 2013; 13: 44.
- Hiroki E, Suzuki F, Akahira J, Nagase S, Ito K, Sugawara J, et al. MicroRNA-34b functions as a potential tumor suppressor in endometrial serous adenocarcinoma. See comment in PubMed Commons below Int J Cancer. 2012; 131: E395-404.
- Ligtenberg MJ, Kuiper RP, Geurts van Kessel A, Hoogerbrugge N. EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. See comment in PubMed Commons below Fam Cancer. 2013; 12: 169-174.
- Tse KY, Liu VW, Chan DW, Chiu PM, Tam KF, Chan KK, et al. Epigenetic alteration of the metallothionein 1E gene in human endometrial carcinomas. See comment in PubMed Commons below Tumour Biol. 2009; 30: 93-99.
- Dehan P, Canon C, Trooskens G, Rehli M, Munaut C, Van Criekinge W, et al. Expression of type 2 orexin receptor in human endometrium and its epigenetic silencing in endometrial cancer. See comment in PubMed Commons below J Clin Endocrinol Metab. 2013; 98: 1549-1557.
- Esteller M, Cordon-Cardo C, Corn PG, Meltzer SJ, Pohar KS, Watkins DN, et al. p14ARF silencing by promoter hypermethylation mediates abnormal intracellular localization of MDM2. See comment in PubMed Commons below Cancer Res. 2001; 61: 2816-2821.
- Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. See comment in PubMed Commons below BMC Cancer. 2006; 6: 212.
- Moreno-Bueno G, Fernandez-Marcos PJ, Collado M, Tendero MJ, Rodriguez-Pinilla SM, Garcia-Cao I, et al. Inactivation of the candidate tumor suppressor par-4 in endometrial cancer. See comment in PubMed Commons below Cancer Res. 2007; 67: 1927-1934.
- Dowdy SC, Gostout BS, Shridhar V, Wu X, Smith DI, Podratz KC, et al. Biallelic methylation and silencing of paternally expressed gene 3 (PEG3) in gynecologic cancer cell lines. See comment in PubMed Commons below Gynecol Oncol. 2005; 99: 126-134.
- Di Domenico M, Santoro A, Ricciardi C, Iaccarino M, Iaccarino S, Freda M, et al. Epigenetic fingerprint in endometrial carcinogenesis: the hypothesis of a uterine field cancerization. Cancer biology & therapy. 2011; 12: 447-457.
- Sasaki M, Kaneuchi M, Fujimoto S, Tanaka Y, Dahiya R. Hypermethylation can selectively silence multiple promoters of steroid receptors in cancers. See comment in PubMed Commons below Mol Cell Endocrinol. 2003; 202: 201-207.
- Li R, Saito T, Tanaka R, Satohisa S, Adachi K, Horie M, et al. Hypermethylation in promoter region of retinoic acid receptor-beta gene and immunohistochemical findings on retinoic acid receptors in carcinogenesis of endometrium. See comment in PubMed Commons below Cancer Lett. 2005; 219: 33-40.
- Banno K, Kisu I, Yanokura M, Masuda K, Kobayashi Y, Ueki A, et al. Endometrial Cancer and Hypermethylation: Regulation of DNA and MicroRNA by Epigenetics. See comment in PubMed Commons below Biochem Res Int. 2012; 2012: 738274.
- Rodriguez-Jimenez FJ, Caldes T. Overexpression of SPARC protein contrasts with its transcriptional silencing by aberrant hypermethylation of SPARC CpG-rich region in endometrial carcinoma. Oncology reports. 2007; 17: 1301-1307.
- Catasus L, Pons C. Promoter hypermethylation contributes to TIMP3 down-regulation in high stage endometrioid endometrial carcinomas. Histopathology. 2013; 62: 632-641.
- Fukami T, Fukuhara H, Kuramochi M, Maruyama T, Isogai K, Sakamoto M, et al. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. See comment in PubMed Commons below Int J Cancer. 2003; 107: 53-59.
- Jiang S, Dowdy SC, Meng XW, Wang Z, Jones MB, Podratz KC, et al. Histone deacetylase inhibitors induce apoptosis in both Type I and Type II endometrial cancer cells. See comment in PubMed Commons below Gynecol Oncol. 2007; 105: 493-500.
- Dvorakova E, Chmelarova M. Methylation analysis of tumor suppressor genes in endometroid carcinoma of endometrium using MS-MLPA. Biomedical papers of the Medical Faculty of the University Palacky, Olomouc, Czechoslovakia. 2013; 157: 298-303.
- Schirmer U, Fiegl H, Pfeifer M, Zeimet AG, Müller-Holzner E, Bode PK, et al. Epigenetic regulation of L1CAM in endometrial carcinoma: comparison to cancer-testis (CT-X) antigens. See comment in PubMed Commons below BMC Cancer. 2013; 13: 156.
- Hu Q, Yu L, Liao QP. [Methylation of matrix metalloproteinases-2 promoter in endometrial cancer invasion]. See comment in PubMed Commons below Beijing Da Xue Xue Bao. 2012; 44: 911-915.
- Bi FF, Li D, Yang Q. Hypomethylation of ETS transcription factor binding sites and upregulation of PARP1 expression in endometrial cancer. See comment in PubMed Commons below Biomed Res Int. 2013; 2013: 946268.
- Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang Y, et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. See comment in PubMed Commons below Nature. 2005; 438: 981-987.
- Xie R, Loose DS, Shipley GL, Xie S, Bassett RL Jr, Broaddus RR, et al. Hypomethylation-induced expression of S100A4 in endometrial carcinoma. See comment in PubMed Commons below Mod Pathol. 2007; 20: 1045-1054.
- Ueda T, Takai N. Apicidin, a novel histone deacetylase inhibitor, has profound anti-growth activity in human endometrial and ovarian cancer cells. International journal of molecular medicine. 2007; 19: 301-308.
- Uchida H, Maruyama T, Ohta K, Ono M, Arase T, Kagami M, et al. Histone deacetylase inhibitor-induced glycodelin enhances the initial step of implantation. See comment in PubMed Commons below Hum Reprod. 2007; 22: 2615-2622.
- Xiong Y, Dowdy SC, Gonzalez Bosquet J, Zhao Y, Eberhardt NL, Podratz KC, et al. Epigenetic-mediated upregulation of progesterone receptor B gene in endometrial cancer cell lines. See comment in PubMed Commons below Gynecol Oncol. 2005; 99: 135-141.