
Research Article
Austin J Clin Pathol. 2017; 4(1): 1044.
MUC1/CD10/ESA Progenitor Cells Immunophenotypes Expression in Breast Cancer and its Relation with Molecular Subtypes
Zanetti JS, Duarte A* and Ribeiro-Silva A
Department of Pathology and Forensic Medicine, Ribeirao Preto Medical School, University of Sao Paulo, Brazil
*Corresponding author: Duarte A, Department of Pathology and Forensic Medicine, Ribeirao Preto Medical School, University of Sao Paulo, Ribeirao Preto, Sao Paulo, Brazil
Received: December 12, 2016; Accepted: January 09, 2017; Published: January 12, 2017
Abstract
Breast carcinomas can be divided in at least four subtypes according to their molecular profile: Luminal A (ER+/HER2-), luminal B (ER+/HER2+), HER2 overexpressing (ER-/HER2+) and basal like (ER-/HER2-). In breast tissues, progenitor’s cells MUC1+/CD10-/ESA+ can differentiate in luminal cells; progenitor’s cells MUC1-/CD10+/ESA- can differentiate in myoepithelial (basal) cells, and progenitors cells MUC1-/CD10+/ESA+ can originate both lineages. The role of these progenitors’ immunophenotypes in breast carcinoma pathogenesis, and its relation with molecular subtypes, still merits further investigation. An immunohistochemical study was performed in a tissue microarray containing 86 samples of breast cancer. We found that the MUC1+/ CD10-/ESA+ immunophenotype is the more frequent progenitor cells phenotype in breast carcinomas, and it correlates with the luminal subtypes.
Keywords: B4reast cancer; Stem cells; Molecular subtypes; ESA
Abbreviations
α-SMA express: α-Smooth Muscle Actin; CD: Cluster of Differentiation; CISH: Chromogenic in Situ Hybridization; CK: Cytokeratin; DAB: Diaminobenzidine; ER: Estrogen Receptor; ESA: Epithelial Specific Antigen; GRB7: Growth factor Receptor Bound Protein 7; H&E: Hematoxylin and Eosin; HER2: Human Epidermal growth factor Receptor 2; MUC1: Mucin 1; PR: Progesterone Receptor; TMA: Tissue Microarray; TP53: Tumor Protein p53
Introduction
Molecular subtypes of breast cancer
Breast carcinomas can be divided in at least four subtypes according to their molecular profiles: luminal A, luminal B, HER-2 overexpressing and basal-like (triple negative phenotype) [1–3]. This classification is a result of molecular characteristics of breast cancer disease. Breast disease progress is a multiple genomic and epigenomic steps. In accordance with Korkola and Gray, breast cancer genomes are deregulated through mutational process and the number of genes deregulated via mutations is less than through other mechanisms [4]. These genetic alterations are, in most part, an amplification of a number of oncogenes and an inactivation of a number of tumor suppressor genes [5].
Luminal phenotype is the most common molecular profile and it is characterized by positivity for estrogen receptors. These carcinomas express cytokeratins 7, 8, 18 and 19 besides some genes related with estrogen receptors activation such as cyclin D1 [1]. Luminal immunophenotype carcinomas can be divided in at least two subtypes: luminal A and luminal B. The subtype luminal A is associated to a larger variety of genes related to estrogen receptor and fewer proliferative genes than subtype B [2]. On the other hand, luminal B carcinomas are more often with high histological grade, high proliferation grade and worst prognostic than luminal A [6]. HER2 phenotype is mainly characterized by HER2/neu, GRB7, and TRAP100 gene expressions. These tumors show high mutation grade on TP53 gene and usually are associated with poor prognosis, independent of grade. Besides, HER2 immunophenotype shows increased mitotic activity, high degree of nuclear pleomorphism and lymph node positivity [7]. The basal phenotype is characterized by cytokeratins with high molecular weight such as cytokeratins 5 and 17 (CK5 and CK17) [8]. Usually, these tumors are negative for estrogen and progesterone receptors and are related with p53, P-cadherin and epithelial growth factor receptor [6]. Basal tumors have a high mitotic grade and pronounced nuclear pleomorphism, resulting in a high Nottingham grade [9]. This kind of tumors represent 17- 37% of all breast carcinomas [10].
Stem cells and progenitors cells
Stem cells are cells able to self-renew and to generate daughter cells that can generate different cell lineages. Self-renewal and differentiation are recognized features of cancer stem cells, as well as high migration capacity and drug resistance [11]. In a mature tissue, these cells have asymmetric divisions in which one or both of the daughter cells are stem cells that retain the same developmental potential as the mother cell [12]. The cells in the intermediate state between stem cell and the terminally differentiated cell are called progenitors or transient cells. The most traditional theory about cancer pathogenesis is that cancer is originated from a single cell that acquires the capacity to propagate indefinitely. Some of these cells share some features with normal adult stem cells, so they are called tumoral stem cells [13]. Because of clonal expansion, most of these cells can be differentiated into other cellular types, and they lose their stem cell abilities. Despite that, a part of these cells remains in their original state, and some authors believe that are these cells that sustain the tumoral growth and determine the therapeutic resistance [14]. One of the major challenges in current oncology is to identify and to study these tumoral stem cells [15]. Some human mammary stem cells have already been identified, and it had led to the hypothesis that these cells may be the origin of breast cancer [13]. Tumoral cells with stem cells features have already been identified in several cancers such as brain, prostate and skin [16,17]. In breast cancer, the most studied neoplastic cells with stem cells features are those with strong CD44 expression and CD24 low or missing (CD44+ CD24- immunophenotype) [18]. These cells are one hundred more tumorigenic than cells without this immunophenotype [16]. In addition, are many markers related to stem progenitor’s cells of the breast, such as MUC1, CD10, and ESA [19]. MUC1 is a protein membrane of mucin family that sometimes, in breast cancer, can be finded in aberrant intracellular localization with glycosylation changes and can be associated with breast carcinoma development [20]. MUC1 aberrant expression or overexpression term, is characterized by accumulate of this glycoprotein in the hole cell membrane, cytoplasm and sometimes in the nucleus [21]. Though this overexpression is associated with poor prognosis and amplified risk of metastasis [22]. Common acute lymphoblastic leukemia antigen (CD10) is a zinc-dependent cell-surface metallopeptidase that can be founded in higher expression in some kind of cancers such as metastatic breast carcinoma and melanoma [23]. CD10 is involved in mammary gland growth and also it can be expressed by normal cells such as fetal liver, bone marrow, spleen, and brain [24–26]. In solid tumors, including breast, CD10 expression is correlated with poor prognosis (mainly if this was located in stroma) [27].
Epithelial Specific Antigen (ESA) has been considered a tumor cell marker in breast, prostate and pancreas carcinoma [19,28]. ESA is a transmembrane protein that is mainly expressed in the basolateral domain of epithelial cell membranes. It belongs to type 1 membrane glycoprotein family [29]. Its over-expression has been reported in breast cancer and retinoblastoma [30]. ESA is involved in metastasis of adenocarcinomas, including breast cancer, and can play an essential role in oncogenic signaling pathways through its proteolysis and intracellular domain translocation into the nucleus [31].
The association with these 3 markers (MUC1, CD10 and ESA) is very important in progenitor cells study in breast cancer. In breast, progenitors cells MUC1+/CD10-/ESA+ can differentiate in luminal cells (that express ESA and some other antigens types); progenitors cells MUC1-/CD10+/ESA- can differentiate in myoepithelial cells (that express α- smooth muscle actin (α-SMA), vimentin and α6 integrin), and progenitors cells MUC1-/CD10+/ESA+, can originate both lineages [28].
The role of these progenitors immunophenotypes in breast carcinoma pathogenesis still merits further investigation. In the present study, we verified these immunophenotypes expression (MUC1+/CD10-/ESA+, MUC1-/CD10+/ESA- and MUC1-/CD10+/ ESA+) in breast carcinomas and we correlated their expressions with luminal A, luminal B, HER2+ and basal immunophenotypes. Moreover, we correlated these immunophenotypes with classical prognostics factors in breast pathology.
Materials and Methods
Patients
The protocol used in this study followed the ethical guidelines of the 1975 Declaration of Helsinki, and the local Ethics Committee approved it. Eighty-six samples of invasive breast ductal carcinomas diagnosed between 1990 and 2004 were retrieved from the files of the Department of Pathology of Ribeirao Preto Medical School. The following information was extracted from the medical files: age, menopausal status, tumor size, metastasis to regional lymph nodes, recurrence, distant metastasis, death. The local Ethics Committee authorized the retrieval of these clinical data. The patients selection was based on the histopathologic diagnosis. For each case, all available Hematoxylin and Eosin (H&E) stained sections were reviewed to confirm the diagnosis of invasive ductal carcinoma and to select a representative area for performing Tissue Microarray (TMA) block. The cases were graded according to the current guidelines of the Scarff-Bloom & Richardson grading system modified by Elston & Ellis [32]. None of the patients had received any treatment before the diagnostic biopsy procedure.
Tissue Microarray (TMA)
The H&E slides were reviewed by an experienced pathologist to delimit the most significant tumor area in each donor paraffin block. This region was then selected to construct a Tissue Microarray (TMA) paraffin block. For TMA block construction, core biopsies (diameter, 3 mm) were punched from the selected regions of each of the 86 donor paraffin blocks and arrayed into a new recipient paraffin block using a Manual Tissue Arrayed I (Beecher Instruments, Silver Spring, USA). Five-μm thick sections were cut from TMA paraffin block using the Paraffin tape-transfer system (Instrumedics, Saint Louis, USA). One section was stained with H&E to confirm the presence of the tumor by light microscopy.
Immuno histochemistry
All tissue samples had been fixed in 4% neutral formalin and embedded in paraffin. Immunohistochemical staining was performed using the Biocare Medical Mach 4 Universal Polymer Detection (Concord, California, USA). The protocol used was described elsewhere [33]. The dilution and source of the primary antibodies used in this study were: MUC1 (1:50, clone 695, Biocare Medical, Concord, California, USA); CD10 (1:50, clone 56C6, Santa Cruz); ESA (1:50, clone H-90, Santa Cruz), estrogen receptor (ER) (1:100 clone 6F11, Novocastra, Newcastle upon Tyne, UK), progesterone receptor (PR) (1:100, clone 16, Novocastra, Newcastle upon Tyne, UK), Ki67 (1:100, clone MM1, Novocastra, Newcastle upon Tyne, UK), C-erbB2 (1:100, clone CB11, Novocastra, Newcastle upon Tyne, UK), and p53 (1:50, clone DO-7, Novocastra, Newcastle upon Tyne, UK). Dilutions for these antibodies were determined for 5 μm thick sections. The reaction was developed with Diaminobenzidine (DAB) followed by hematoxylin counterstaining.
According to the literature, the immuno histochemistry study (1+) when the positive cells were 5 to 10%; medium positive (2+) was evaluated as follows: the cases in which > 20% of neoplastic cells stained for Ki67 (nuclear staining) were considered highly proliferative [34]; the cases were interpreted as ER, PR and p53 positives if more than 10% of the neoplastic cells showed nuclear staining [32]. For immuno histochemistry positive control for MUC1, CD10 and ESA we used healthy breast tissue. Normal expression of MUC1 is in apical membrane, ESA is founded in basolateral surface and CD10 is founded in stroma and lateral membrane of myoepithelial cells. Cases of invasive ductal carcinoma previously known to be positive for Ki67, ER, PR, p53, C-erbB2, were used as positive controls. Negative controls for immunostaining were done by omission of the primary antibody. Cut-off for MUC1 analysis was the following: negative score when the positive cells for MUC1 were 0 to 5%; weakly positive when the positive cells were 10 to 50% with cytoplasmic expressions, and diffusely positive or superexpression (3+) when the cells stained were more than 50% with cytoplasmic and nucleus expressions [35]. For CD10 cut-off we considered: negative when there were not stained cells, weakly positive when there were less than 30% stained cells and strongly positive when we visualized more than 30% of immunostaining cells [27]. ESA cut-off used was as follows: score 0 for negative staining in all cells; score 1+ for weakly positive or focally positive membrane staining in less than 10% of the cells; score 2+ for moderately positive staining in 10% to 50% of the cells; and score 3+ for strongly positive staining above 50% of the cells [36].
CISH “Chromogenic In Situ Hybridization”
HER-2 immuno expression was scored according to American Society of Clinical Oncology/ College of American Pathologists guideline. For statistical purposes, carcinomas with score 0 or 1+ were considered negative; and carcinomas with score 3+ were considered positive. Cases with HER2scored as 2+ were submitted to Chromogenic in Situ Hybridization (CISH) with ZytoDot 2C SPEC HER2/CEN 17 probe kit (Zytovision, Bremerhaven, Germany). HER2 was considered amplified when the HER2/CEN 17 ratio was ≥ two on average for 60 cells [34,35,37].
Statistical analysis
Statistical analyses and tests were performed with the commercially available PASW Statistics 17.0 software (Chicago, IL, USA). The relations between MUC1/CD10/ESA expression, immunohistochemical findings, and clinicopathologic features were tested with cross tables applying the χ2 (three or more variables) or Fisher tests (two variables), and all tests were 2-tailed. A p-value of <0.05 was considered significant.
Molecular profiles
Luminal A subtype were considered the cases ER positive and HER2 negative, luminal B subtype were the cases ER positive and HER2 positive [9]. HER2 subtype was considered the cases HER2 positive, and ER were negative [9,38]. The basal subtype were the cases negative for both ER and HER2 [39,40].
Results
Molecular profiles and progenitors cells
Among the 86 previously selected patients, we were able to determine the molecular profiles in 69 of them. We had 06 patients with basal phenotype (7.0%), 39 with luminal A phenotype (45.3%), 16 with luminal B subtype (18.6%), 08 patients with the HER-2 profile (9.3%) and 17 patients in which we were unable to determine the molecular profile for technical reasons (19.8%). In accordance with profiles for progenitor cells MUC1+/CD10-/ESA+, MUC1-/CD10+/ ESA- and MUC1-/CD10+/ESA+ we found 47 patients with MUC1+/ CD10-/ESA+ immunophenotype (54.6%), 18 cases for MUC1-/ CD10+/ESA- (20,9%) and finally, 04 cases for MUC1-/CD10+/ ESA+ (4,6%). As observed in (Table 1), the MUC1+/CD10-/ESA+ immunophenotype is the more frequent progenitor cells phenotype, and it correlates with the luminal subtype. (Figures 1 and 2) show an example of ESA and MUC1 immuno histochemistry marker for progenitor cells.
MUC1+/CD10-/ESA+
MUC1-/CD10+/ESA-
MUC1-/CD10+/ESA+
p
Molecular profiles
0,001
Basal
3
2
1
Luminal A
27
11
1
Luminal B
14
1
1
HER2
3
4
1
Total n
47
18
4
Table 1: Molecular profiles with progenitor cells: MUC1+/CD10-/ESA+, MUC1-/ CD10+/ESA- and MUC1-/CD10+/ESA+ profiles.
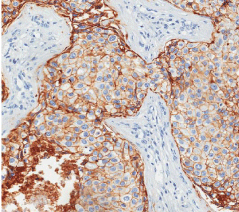
Figure 1: Immunohistochemical assay of breast carcinoma cells for positive
ESA, the magnification of 400 X.
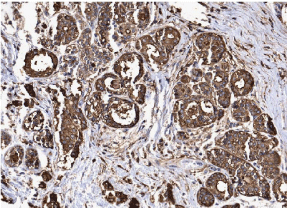
Figure 2: Immunohistochemical assay of breast carcinoma cells for positive
MUC1, the magnification of 200X.
Progenitors cells and clinic-pathological features
Our study showed patients with a 55 median age (range 25- 85 years), 37 patients were less than 50 years, and 49 were more than 50 years. Thirty-two patients were premenopausal, and 54 were posmenopausal. Tumors sizes ranging from 25- 180 mm. Sixteen patients had tumors measuring less than 20 mm, while 40 patients had tumors between 20- 50 mm and 30 with larger than 50 mm. Fortythree patients were lymph node negative, and 43 patients had positive lymph nodes. The tumors were classified as grade I (32 [37.2%]), grade II (44 [51.2%]) and grade III (10 [11.6%]). In agreement with Clinical Staging System, created by the American Joint Committee on Cancer [39], Seven patients were stage I, 27 and 11 were stage IIa and IIb respectively, 15 and 21 patients were stage IIIa and IIIb respectively and finally, 05 were stage IV. Distant metastasis occurred in 24 patients, and we observed 28 deaths. Progenitors cells profiles did not correlate with tumor size, pathological stage, age, menstrual status, nodal status, and tumor grade. All of the clinical and immunohistochemical findings are shown in (Tables 2 and 3).
MUC1+/CD10-/ESA+
MUC1-/CD10+/ESA-
MUC1-/CD10+/ESA+
p
Age (years)
0,305
<50
19
17
1
>50
29
16
4
Menstrual status
0,082
Pre-menopausal
13
17
2
Pos-menopausal
35
16
3
Tumor size (mm)
0,245
<20
11
4
1
20-50
19
20
1
>50
18
9
3
Tumoral grade
0,721
I
17
12
3
II
24
18
2
III
7
3
0
Lymph node
0,164
Positive
26
13
4
Negative
22
20
1
Staging
0,595
I
5
2
0
IIa
15
11
1
IIb
6
3
2
IIIa
8
6
1
IIIb
11
10
0
IV
3
1
1
Recurrence
0,101
Yes
9
1
1
No
39
32
4
Metastasis
0,876
Yes
13
10
1
No
35
23
4
Death
0,736
Yes
17
10
1
No
31
23
4
Total n
48
33
5
Table 2: Progenitors cells profiles and classical clinical pathological factors.
MUC1+/CD10-/ESA+
MUC1-/CD10+/ESA-
MUC1-/CD10+/ESA+
p
ER
0,001
Positive
41
12
2
Negative
7
21
3
PR
0,05
Positive
35
16
2
Negative
13
17
3
Ki67
0,274
Positive
21
14
4
Negative
27
19
1
HER2
0,112
Positive
17
5
2
Negative
37
28
3
p53
0,824
Positive
33
24
8
Negative
15
9
2
Total n
48
33
5
Table 3: Immunohistochemistry founds and relation with progenitors cells.
Discussion
The present study aimed to contribute to understanding the role of MUC1+/CD10-/ESA- and MUC1-/CD10+/ESA- immunophenotypes in breast cancer. We applied immuno histochemistry technique for the identification of these two immunophenotypes in paraffin-embedded tissue sections of patients with breast cancer to determine whether there is any relationship between molecular profiles (basal, luminal A, luminal B and HER2) and classical immuno histochemistry markers as hormone receptors and regulators of cell cycle. The mammary epithelium is composed of two lineages of epithelial cells in which they are luminal and myoepitelial cells. Luminal and myoepithelial cells can be identified by expression of specific proteins. For example, luminal cells can be identified by their expression of ESA and some cytokeratins 7, 8, 18 and 19 and myoepithelial cells express α-smooth muscle actin (α-SMA), α6 integrin, vimentin and CD10 expressions (among others markers). As mentioned before, in breast tissues, progenitors cells MUC1+/CD10-/ESA+ can differentiate in luminal cells, progenitors cells MUC1-/CD10+/ESA- can differentiate in myoepithelial cells and progenitors cells MUC1-/CD10+/ESA+, can originate both lineages. We noticed there are a significant relation of MUC1+/CD10-/ESA+ and MUC1-/CD10+/ESA- with hormone receptors ER and PR. But there are more patients ER+ in MUC1+/ CD10-/ESA+ than in MUC1-/CD10+/ESA-. Usually ER is expressed in 10% to 30% of luminal cells but not in myoepitelial ones [41]. Other point in this discussion is related with the localization of ER in mammary gland. It is localized in luminal epithelial cells [19] and maybe due to MUC1+/CD10-/ESA+ differentiated in luminal cells they express higher ER. Besides, there are more cases positive for p53 in MUC1+/CD10-/ESA+ even though, this result is not statistically significant. P53 positivity in immuno histochemistry means that in the samples, there is p53 inactive indicating some kind of problem in cellular cycle. About the same kind of immunoprofile MUC1+/ CD10-/ESA+ this one is most frequently seen in luminal (A and B) profile and this kind of immuoprofile of progenitor cells may appear with most frequency in women with more than 50 years, in menopausal status. Also, recurrence, metastasis and death seem to be less frequency in this kind of profile.
MUC1+/CD10-/ESA+ profile has an balanced distribution among age, menstrual status, tumoral grade, metastasis and death, but seems to be a better prognostic in relation with recurrence (there are 32 cases with no recurrence against 1). Both immunoprofile MUC1+/CD10-/ESA+ and MUC1-/CD10+/ESA- are most frequency than MUC1-/CD10+/ESA+. In this patients samples we found a high expression of luminal molecular subtype (specifically, luminal A). Basal subtype had almost the same expression in both immuno histochemistry profile while luminal molecular subtype seems to show a better expression on MUC1+/CD10-/ESA+ profiles suggesting a relation with this type of molecular subtype with this progenitor differentiation (even though, luminal B seems to have no difference in both immuno histochemistry profiles). As for HER2 subtype, we cannot affirm whether this one has more or less expression in which or those immuno histochemistry profiles due to a low quantity of patients expressing this profile. Immuno histochemistry for progenitor’s cells identification is a quick and efficient test to study the possible role of tumoral progression in breast cancer. Our finds show that luminal immunophenotype is commonly associated with MUC1+/CD10-/ESA+ progenitor phenotype. There are already some studies in the literature indicating that MUC1 positivity is associated with ESA and our findings corroborate with these studies [28].
Conclusion
The understanding of breast cancer behavior and its subtypes has been changed with the advent of widespread screening programs and the systematic use of adjuvant hormonal therapy are resulting a major impact in outcome and decreasing breast cancer mortality. Due to this, studies like ours that show breast cancer behavior have been considering an important tool to early diagnosis, classification of the tumor and individualize patient’s treatment. Despite of genomic technologies the most common assay used in diagnostic routine is immuno histochemistry. With this article we show an easy and functional way to correlate the diagnosis with poor prognosis and histological subtype to help in diagnosis routine. Here we noticed that MUC1+/CD10-/ESA+ immuno histochemistry profile has several characteristics that suggest some genetics alterations and this profile is most frequency than MUC1-/CD10+/ESA-. Besides, we hope this work may contribute to others studies with progenitors cells and once we can understand what kind of influence that these cells may be cause in tumors differentiation. Thereby, these studies will be use in new therapies that prevent self-renewal of breast cancer cells.
References
- Perou CM, Sorlie T, Eisen MB, Van De Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000; 17: 747–752.
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003; 100: 8418-8423.
- Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004; 15: 5367-5374.
- Korkola J, Gray JW. Breast cancer genomes form and function. Curr Opin Genet Dev. 2010; 20: 2-18.
- Van De Vijver MJ. Genetic alterations in breast cancer. Curr Diagnostic Pathol. 2000; 6: 271-281.
- Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010; 4: 192-208.
- Hanby AM. Aspects of molecular phenotype and its correlations with breast cancer behaviour and taxonomy. Br J Cancer. 2005; 92: 613-617.
- Rijn M, Van De, Perou CM, Tibshirani R, Haas P, Kallioniemi O, et al. Outcome. 2002; 161: 1991-1996.
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes and survival in the Carolina Breast Cancer Study. JAMA. 2006; 295: 2492-2502.
- Foulkes WD. Germline BRCA. Mutations and a Basal Epithelial Phenotype in Breast Cancer. Cancer Spectrum Knowl Environ. 2003; 95:1482-1485.
- Burkert J, Wright NA, Alison MR. Stem cells and cancer: an intimate relationship. J Pathol. 2006; 209: 287-297.
- Pine SR. Asymmetric cell division and template DNA co-segregation in cancer stem cells. 2014; 4: 1-6.
- Cariati M, Purushotham AD. Stem cells and breast cancer. Histopathology. 2008; 52: 99-107.
- Phillips TM, McBride WH, Pajonk F. The response of CD24 (-/low)/CD44+ breast cancer-initiating cells to radiation. J Natl Cancer Inst. 2006; 98: 1777-1785.
- Ponti D, Zaffaroni N, Capelli C, Daidone MG. Breast cancer stem cells: an overview. Eur J Cancer. 2006; 42: 1219-1224.
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003; 100: 3983-3988.
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005; 65: 10946-10951.
- Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005; 11: 1154-1159.
- Stingl J. Detection and analysis of mammary gland stem cells. J Pathol. 2009; 217: 229-241.
- Bon GG, Van Kamp GJ, Verstraeten RA, Von Mensdorff-Pouilly S, Hilgers J, Kenemans P. Quantification of MUC1 in breast cancer patients. A method comparison study. Eur J Obstet Gynecol Reprod Biol. 1999; 83: 67-75.
- Hattrup CL, Gendler SJ. MUC1 alters oncogenic events and transcription in human breast cancer cells. Breast Cancer Res. 2006; 8: 37.
- Rahn JJ, Dabbagh L, Pasdar M, Hugh JC. The importance of MUC1 cellular localization in patients with breast carcinoma: an immunohistologic study of 71 patients and review of the literature. Cancer. 2001; 91: 1973-1982.
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006; 3: 187-197.
- Moritani S, Kushima R, Sugihara H, Bamba M, Kobayashi TK, Hattori T. Availability of CD10 immunohistochemistry as a marker of breast myoepithelial cells on paraffin sections. Mod Pathol. 2002; 15: 397-405.
- Tse GMK, Tan P-H, Lui PCW, Gilks CB, Poon CSP, Ma TKF, et al. The role of immunohistochemistry for smooth-muscle actin, p63, CD10 and cytokeratin 14 in the differential diagnosis of papillary lesions of the breast. J Clin Pathol. 2007; 60: 315-320.
- Tse GMK, Tsang a KH, Putti TC, Scolyer RA, Lui PCW, Law BKB, et al. Stromal CD10 expression in mammary fibroadenomas and phyllodes tumours. J Clin Pathol. 2005; 58: 185-189.
- Makretsov NA, Hayes M, Carter BA, Dabiri S, Gilks CB, Huntsman DG. Stromal CD10 expression in invasive breast carcinoma correlates with poor prognosis, estrogen receptor negativity, and high grade. Mod Pathol [Internet]. 2007; 20: 84-89.
- Stingl J, Eaves CJ, Kuusk U, Emerman JT. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998; 63: 201-213.
- Lu X, Li H, Xu K, Nesland JM, Suo Z. MUC-1-/ESA+ progenitor cells in normal, benign and malignant human breast epithelial cells. Histol Histopathol. 2009; 24: 1381-1390.
- Baeuerle P, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007; 96: 417-423.
- Schnell U, Kuipers J, Giepmans B. EpCAM proteolysis: new fragments with distinct functions? Biosci Rep. 2013; 33: 321-332.
- Fitzgibbons PL, Page DL, Weaver D, Thor a D, Allred DC, Clark GM, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement. Arch Pathol Lab Med. 2000; 124: 966-978.
- Ribeiro-Silva A, Moutinho MAK, Moura HB De, Vale FR Do, Zucoloto S. Expression of checkpoint kinase 2 in breast carcinomas: correlation with key regulators of tumor cell proliferation, angiogenesis and survival. Histol Histopathol. 2006; 21: 373-382.
- Cheang MCU, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009; 101: 736-750.
- Sangoi AR, Higgins JP, Rouse R V, Schneider AG, McKenney JK. Immunohistochemical comparison of MUC1, CA125, and Her2Neu in invasive micropapillary carcinoma of the urinary tract and typical invasive urothelial carcinoma with retraction artifact. Mod Pathol [Internet]. 2009; 22: 660-667.
- Qun LIU, Ji-guang LI, Xin-yu Z, Feng JIN, Hui-ting D, Cscs T. Expression of CD133, PAX2, ESA and GPR30 in invasive ductal breast carcinomas. Chin Med J (Engl). 2009; 122: 2763-2769.
- Garcia-Caballero T, Grabau D, Green AR, Gregory J, Schad A, Kohlwes E, et al. Determination of HER2 amplification in primary breast cancer using dual-colour chromogenic in situ hybridization is comparable to fluorescence in situ hybridization: a European multicentre study involving 168 specimens. Histopathology. 2010; 56: 472-480.
- Blows FM, Driver KE, Schmidt MK, Broeks A, Van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010; 7: e1000279.
- Tan EY, Yan M, Campo L, Han C, Takano E, Turley H, et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer [Internet]. 2009; 100: 405-411.
- Rakha EA, El-Sayed ME, Green a R, Paish EC, Lee a HS, Ellis IO. Breast carcinoma with basal differentiation: a proposal for pathology definition based on basal cytokeratin expression. Histopathology. 2007; 50: 434-438.
- Clayton H, Titley I, Vivanco M DM. Growth and differentiation of progenitor/stem cells derived from the human mammary gland. Exp Cell Res. 2004; 297: 444-460.