Research Article
Austin J Comput Biol Bioinform. 2014;1(1): 9.
The Association of Set3 Complexes with Nucleosomes and whole Genome in Saccharomyces Cerevisiae
Tung SY1, Tsai HC2, Shen HH2, Tsai SP1, Lee SP1, Hsiao SP2, Chen FJ2 and Liou GG2,3,*
1Insitute of Molecular Biology, Academia Sinica, Taiwan
2Institute of Molecular and Genomic Medicine, National Health Research Institutes, Taiwan
3Graduate Institute of Basic Medical Science, China Medical University, Taiwan
*Corresponding author: Liou GG, Institute of Molecular and Genomic Medicine, National Health Research Institutes, 35 Keyan Road, Jhunan Town, Miaoli county 35053, Taiwan
Received: May 19, 2014; Accepted: June 17, 2014; Published: June 22, 2014
Abstract
The Set3 Complex (SET3C) is involved in several biological processes such as transcription. SET3C is capable of function not only in the repression of gene expression but also in the activation. To characterize the molecular property of SET3C in more detail, here, we used a typical TAP tag method and succeed to directly purify a native Set3-TAP complex from yeast and so did Snt1-TAP complex. Both the components of Set3-TAP complex and of Snt1-TAP complex were same as the reported model of SET3C, including the proteins Snt1, Hos4, Set3, Sif2, Hos2, Hst1 and Cpr1. Furthermore, we also demonstrated that the SET3C was able to associate with nucleosome by several biophysical methods. And through the Chromatin Immunoprecipitation (ChIP) on chip assay, we also analyzed 4 data sets of the genome-wide localization of SET3C to identify 590 major association gene regions of SET3C. In summary the positive and negative role on the modulation of gene expression could consist with the association of SET3C with native nucleosomes and also agree with the exploration of dynamic global genome association of SET3C.
Keywords: ChIP; Genome-wide; Protein-protein interaction; Nucleosome; Set3; SET3C
Abbreviations
ChIP: Chromatin Immunoprecipitation; Co-IP: Co-immunoprecipitation; EM: Electron Microscopy; ITC: Isothermal Titration Calorimetry; SET3C: Set3 Complex; SPR: Surface Plasmon Resonance; TAP: Tandem Affinity Purification
Introduction
In the cell, the nucleosome is the basic organizational unit of chromatin and consists of approximate 146 base pairs of DNA wrapped twice around an octamer core of histones (H2A, H2B, H3 and H4) [1- 3]. The epigenetic modifications of nucleosome, accompanied with histone association proteins, organize the genome into distinct states, which are referred to as heterochromatin and euchromatin [4-6]. Euchromatin is known as the transcriptionally active portion of the genome, whereas heterochromatin is defined as the transcriptionally less-active condensed chromosome regions. Chromatin displays epigenetic inheritance, in that changes in its structure can pass to the next generation independently of the DNA sequence itself. It is now clear that the modifications of histones, particularly at their N-termini, including acetylation, methylation, phosphorylation, sumoylation, ubiquitination, deimination, isomerization, glycosylation, and ADP-ribosylation, modulate the activity of many genes by modifying the affinity between histones and DNA, and between histones and their associated proteins. These epigenetic modifications play an important role in multiple cellular processes including DNA repair, genome stability, and gene expression [4,7-9]. Although there has been significant progress in identifying and understanding the molecular mechanisms of these epigenetic modifications, a lot of them still remain to be more characterized.
The Set3 Complex (SET3C) has been discovered by co-purification experiments of tagged proteins that had generated a model of SET3C, including Set3, Sif2, Snt1, Hos4, Cpr1, and two histone deacetylases, Hos2 and Hst1 [10]. Set3 has a highly sequence similarity to mammalian MLL5. There is a similar human corepressor complex, Silencing Mediator for Retinoid and Thyroid receptor (SMRT) [11,12]. Both SET3C and SMRT complexes have some components containing SET domain, SANT domain, histone deacetylase activity, and WD repeat domain. The SET domain is suggested may be involved in the histone methylation. SET3C is possible the first discovered complex that related to both methylation and deacetylation of histone. For epigenetic point of view, specific histone methylation and deacetylation are much positively correlation with gene expression [13-15].
The SET3C has been reported to be involved in several biological processes, such as meiosis-specific repression of sporulation, promotion of Ty1 retrotransposons integrating at tRNA genes and signal transduction of secretory stress through the Mpk1p cell integrity pathway [10,16,17]. The SET3C has been also reported to have functions of repressive and activating of gene expression. A model for the positive transcription effect of Set3 has been proposed. Set1 makes H3K4 dimethylation that recruits SET3C to promoter-proximal regions. Then, Hos2 and Hst1 of SET3C make nucleosome deacetylation near 5' regions of genes. The deacetylation of nucleosomes, located at downstream of the promoter, may promote efficient elongation of RNA transcription by RNA polymerase II [18]. The PHD finger of Set3 is able to bind to methylated H3K4 in vitro [19]. However, SET3C has been also proposed to be recruited to transcribed regions by interaction with the phosphorylated CTD of RNA polymerase II but not required H3K4me2 for the further histones deacetylation [20]. Anyhow, SET3C negatively regulates meiotic genes and is required for positive induction effect of the GAL1 and INO1 genes expression [10,21]. SET3C not only processes deacetylation of histones in 5' transcribed regions [18] but also is capable of modulating the kinetics of many transcriptional responses for the changes of carbon source. And the major target genes of Set3 overlap with noncoding RNA transcription [22-25].
To investigate more detail of the SET3C property, here, we reported that we succeed to directly purify a native SET3C from yeast via the typical TAP tag method. We also showed the evidences of the interaction of SET3C with nucleosomes by several distinct methods. Furthermore, through the Chromatin Immunoprecipitation (ChIP) on chip assay, we identified the major association genes of SET3C by analysis of the multiple data sets of the genome-wide localization of SET3C.
Materials and Methods
Yeast Strains
The yeast strains used for this study were W303-1a (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1). GLY002 (Snt1-TAP TRP1. W303-1a), GLY003 (Set3-TAP TRP1. W303-1a). Strains DMY3392 (HTA2-TAP-K.l.-TRP1, W303-1a) was previously described [26].
Protein purification, gel electrophoresis, coimmunoprecipitation (Co-IP), western blot
The purification methods of H2A-TAP, Set3-TAP and Snt1- TAP complexes were same as described as previously [26,27]. Protein samples were separated on SDS-PAGE and were stained by Coomassie brilliant blue R520 to visualize protein bands. Co-IP was based on a method described previously [26]. Briefly, 20 g of frozen cell pellets of SET3C were lysed by bead beating (Biospec) in an equal volume of lysis buffer (50mM HEPES-KOH [pH 7.6]; 10mM Mg(OAC)2; 5mM EGTA; 0.1mM EDTA; 150mM KCl; 0.2% NP-40; 5% glycerol; 2mM phenylmethylsulfonyl fluoride; 1 tablet of protease inhibiter cocktail; and 1mM benzamidine). The lysate was bound to 100 μl IgG sepharose bead (GE healthcare) for 2-4 hours. The beads were washed five times with 1 ml of wash buffer A (50mM Tris-HCl [pH 7.5], 150mM NaCl, 0.05% NP-40). Then, 50ul of 1mM purified nucleosome was added to the bead to incubate with SET3C for 2-4 hours. The beads were washed five times with 1 ml of wash buffer B (50mM Tris-HCl [pH 8.0], 150mM NaCl, 0.05% NP-40). The bound sample beads were eluted by incubation with 10 ul of home-made TEV protease for overnight at 4oC. Western blotting was performed using Immobilon Western detection system (Millipore).
Electron microscopy (EM)
The sample negative-staining of EM was as described as previously [28] with some modifications. Briefly, 1 drop of 15 μl sample was adsorbed to a glow-discharged 200-mesh copper grid covered with carbon-coated collodion film, washed with 3 drops of 20μl distilled water, and stained with 2 drops of 20μl 0.75% uranyl formate. Samples were examined with an FEI Tecnai T12, a Hitachi H7650 or a Jeol 2100F electron microscope.
BIAcore surface plasmon resonance analysis
Real time protein-protein interactions were examined using a BIAcore 2000 instrument (BIAcore). SET3C, nucleosome or BSA was individually immobilized on different flow cell of a CM5 sensor chip using an amine-coupling kit (BIAcore). The experimental procedures for interaction assays and data analysis were previously described [29].
Isothermal titration calorimetry (ITC)
ITC measurement was performed on a VP-ITC system (MicroCal, Inc.), using the VP Viewer software for data acquisition and instrument control. All protein samples were extensively dialyzed against 20mM Hepes-KOH pH 8.0, 300mM KCl, 1mM MgCl2, 50 μM NAD. All samples and buffers were filtered and degassed before use. The concentration of nucleosome in the injection syringe was 50 μM and the concentration of SET3C in the reaction cell was 5 μM. In a typical experiment, after an initial 1.5 μl injection, 29 aliquots of 10 μl were titrated at 4 minute intervals from the syringe into the 1.4 ml sample cell, maintained at 20°C and stirred at a constant rate of 270 rpm to ensure rapid mixing. The titration curves of binding isotherm data were analyzed using the MicroCal Origin software package, assuming one set of sites to obtain the binding constant (K), enthalpy (ΔH) and entropy (ΔS) with their standard error for fitting to the data.
Chromatin immunoprecipitation (ChIP) on Chip
The ChIP on chip assays were as described as previously [30] with some modifications. Briefly, the precipitation chromatin DNA fragments of Set3-TAP or Snt1-TAP associated by an IgG sepharose bead (GE healthcare) were gotten, then, following the Nimble Gen standard protocols instructions, samples were processed from the steps of post IP, of ligation mediated PCR (LM-PCR), of labelling with either cy5 or cy3, of hybridization with S. cerevisiae whole genome tiling array chip (NimbleGen (currently Roche)), to the step of scanning chip and analysis of data using the Nimble Scan program and Signal Map software. Data had been deposited in GEO (accession #GSE57916)
Results and Discussion
Association of Set3 complexes with nucleosomes
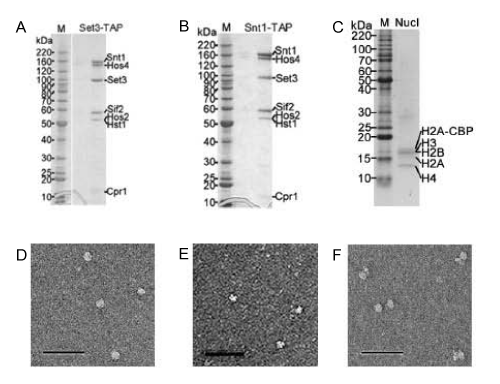
We used Electron Microscopy (EM) examination, Co-immunoprecipitation (Co-IP), BIAcore Surface Plasmon Resonance (SPR) and Isothermal Titration Calorimetry (ITC) approaches to verify whether the Set3 Complexes (SET3C) were able to directly interact with nucleosomes. The native SET3C and nucleosomes were purified from yeast (Figure 1A-C). Using the typical TAP tag strategy, we modified the Set3 and Snt1 genes of Saccharomyces cerevisiae, budding yeast, chromosome as the fusion Set3-TAP and Snt1-TAP genes, respectively. And through a well developed TAP tag purification method, we succeed to directly purify a native Set3-TAP and Snt1-TAP complexes from yeast. The purified Set3-TAP and Snt1-TAP complexes were shown as figure 1 with a denaturing 4 ~20 % gradient SDS PAGE. According the mobility of each protein bands, both our Set3-TAP and Snt1-TAP complexes were same as the reported SET3C, and contained Snt1, Hos4, Set3, Sif2, Hos2, Hst1, and Cpr1. They had also been confirmed by the Matrix-Assisted Laser Desorption Ionization Time-Of-Flight (MALDI-TOF) mass spectrometer.
Figure 1: Purified native SET3C and nucleosomes. The Purified Set3-TAP complexes (A) and Snt1-TAP complexes (B) were separated by 4~20 % SDSPAGE. The purified nucleosomes (C) were separated by 20 % SDS-PAGE. Gels were stained with Coomassie brilliant blue. The components of complexes were individually indicated. The molecular masses of protein standard (kDa, M) were shown. Electron Micrographs were shown the negatively stained purified Snt1-TAP complex (D), nucleosomes (E) and the particles in the solution of SET3C incubation with nucleosomes (F), respectively. The bar represented 100 nm.
Examination by EM, we were able to directly observe the whole structure of SET3C and the morphological feature of SET3C was detected as a ball shape (Figure 1D). Furthermore, we found a distinct morphological feature of the particle containing in the incubation solution mixed SET3C with nucleosomes. In general, nearly half of the observed particles appeared globular adopted with extra two smaller domains suggesting that association may be also induced a structural rearrangements. However, neither SET3C nor nucleosome particles were detected as the same type particles as above described shape (Figure 1 D-F). It was implied that SET3C was able to association with nucleosomes.
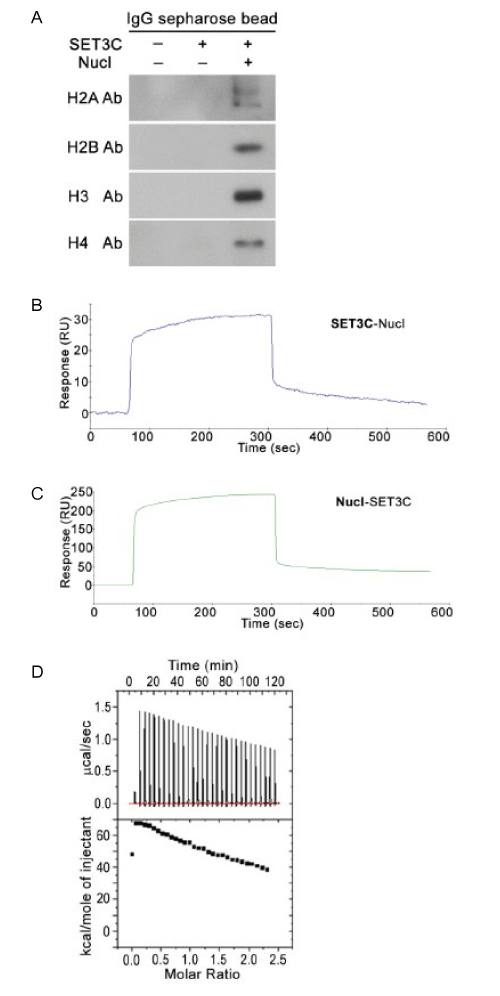
We then used the Immunoprecipitation Western (IP-western) to test whether a native purified nucleosomes was able to be Co-IP with SET3C. We incubated SET3C, from strain expressing Snt1-TAP (or Set3-TAP (data not shown)) immobilized on IgG-Sepharose beads, with purified nucleosomes to allow SET3C association with nucleosomes. Following the removal of unbound nucleosomes by several washes, the possible entire complex of SET3C-nucleosome was eluted by TEV protease. We then used individual histone antibody, H2A. H2B, H3, and H4, to examine whether entire nucleosome was association with SET3C by this Co-IP assay. The results were shown as Figure 2A that all 4 histones were detected. We noticed that there were two detected bands by histone 2A antibody. It was consisted with the protein pattern of our purified nucleosome (Figure 1C) we believed that because the yeast strain for our purified nucleosomes, contained one locus of H2A-TAP gene (HTA2-TAP) and another locus of H2A gene (HTA1) on the genome. However, the results agreed with the association of SET3C with nucleosomes.
Figure 2: Association of SET3C with nucleosomes. The results of IPwestern were shown that nucleosomes (Nucl) were co-immunoprecipitated by SET3C. The 4 core histones were detected by the specific antibody as indicated, respectively (A). Representative Sensorgrams of BIAcore SPR were shown the binding of Nucleosome (Nucl) to immobilized SET3C (B) and the binding of SET3C to immobilized Nucleosome (Nucl) (C). ITC assay measured the association of SET3C with nucleosomes and the obtained data were analyzed by the MicroCal Origin software package (D).
To confirm the directly binding of SET3C to nucleosomes, we performed BIAcore SPR assays. SET3C, nucleosome and control BSA were individual immobilized to a CM5 sensor chip, and then nucleosomes and SET3C were individually used as the analyzers. Binding was observed between SET3C and nucleosome, no matter either SET3C or nucleosome was immobilized on the sensor chip (Figure 2B-C). The on and off rates for the association of SET3C with nucleosomes were not measured, because we were not sure whether entire SET3C (or nucleosome) was still immobilized on the chip after the degeneration washes to disassociate the nucleosome from SET3C (or vice versa). We did not want to make the artificial fixed complex particles before we did the interaction assays. It was ambiguous that the artificial fixation of complex particles might affect the property of their real interactions.
To further measure the association of SET3C with nucleosomes under equilibrium condition, we used ITC assay. In this assay, SET3C (50 μM) was injected into a reaction cell containing nucleosome (5 μM). As shown in Figure 2D, the interaction of SET3C with nucleosome was demonstrated. The calculated affinity of SET3C with nucleosomes was 85.4nM. Furthermore, the interaction of SET3C with nucleosomes exhibited endothermic properties (ΔH = 44.4 kcal/mole) and a positive entropy value (ΔS = 174.2 cal/mole/deg), suggesting that binding may be accompanied by a structural changes. It was agreement with our morphological observation by EM.
Genome-wide localization of Set3 complexes
We applied the ChIP assay in combination with a whole genome tiling array chip to determine the genome-wide association of SET3C. As described as above, we found SET3C could directly bind nucleosome and purified Set3-TAP complexes and Snt1-TAP complexes were same as the SET3C, therefore we performed ChIP-chip experiments using chromatin immunoprecipitation of Set3-TAP and Snt1-TAP and try to map the major genome-wide association of SET3C.
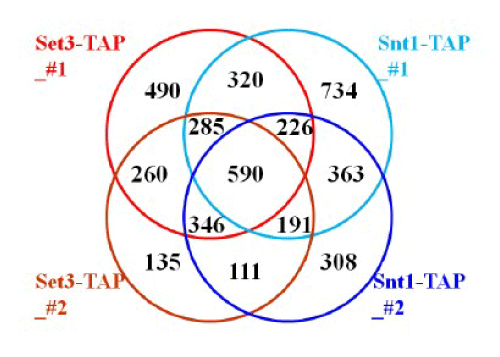
We performed data analysis by calculating and filtering out peaks with False Discovery Rate (FDR) through a suit of NimbleScan software. As expectation, the results of 4 data sets were not totally identical. In general, each data contained around 2000 ~ 2500 association genes (1918, 2125, 2517 and 2648 genes, respectively). Briefly, using the Sir3-TAP strain, according to two distinct biological data sets, we observed that the localization gene number of SET3C was 1481, whereas, according to two distinct biological ChIP on chip data sets of Snt1-TAP, we identified the localization gene number of SET3C as 1360 (Figure 3). In summary, we mapped 590 potential SET3C association genes (Figure 3). The analysis result of Gene Ontology (GO) was shown in Table 1. These 590 SET3C-associated genes were diverse and involved in many biological processes such as transcription from RNA polymerase II promoter (57 out of 590 genes), response to chemical (55 out of 590 genes), carbohydrate metabolic process (39 out of 590 genes), generation of precursor metabolites and energy (23 out of 590 genes), meiotic cell cycle (33 out of 590 genes) and cytoplasmic translation (21 out of 590 genes). At same time, other ontological analysis such as molecular functions (Table S1) and cellular components (Table S2) also showed a similar diversity. However, it remains to be determined whether the SET3C directly play any role in the regulation of these loci.
Figure 3: Cross comparisons of number of genes association with SET3C. The association genes of SET3C were identified from the ChIP on chip assays by analysis of data set with the standard false discovery rate (FDR) calculating and filtering out peaks. The four biological data sets were indicated as Set3-TAP_#1 (red), Set3-TAP_#2 (brown), Snt1-TAP_#1 (light cyan), and Snt1-TAP_#2 (blue), respectively.
Characters in each type of headache
Type
Number
Gender
Age
MRI (T1)
MRI (T2)
Waxy nodule
E.A.
V.D.
Size (mm)
Location
Duration
Surgery
w/o improvement
Recurrence
(%)
(years)
(%)
(%)
(%)
(%)
(%)
(mm)
after surgery
after surgery
Apoplexy
15 (24)
1 ; 14
31.7
hyper 15 (100)
hypo 15 (100)
15 (100)
9 (60)
2 (13)
15.3
frontal 11 (73)
1-6d
11 (73)
0 (0)
1 (7)
temporal 2 (13)
TC 0, TS 11
0 (0)
0 (0)
occipital 1 (7)
retro-bulber 1 (7)
Acute
23 (37)
9 ; 14
34.3
hyper 23 (100)
hypo 18 (78)
6 (26)
5 (28)
3 (17)
14.6
frontal 16 (70)
1w-1m
14 (61)
1 (4)
1 (4)
hyper 5 (22)
2 (9)
3 (60)
1 (20)
retro-bulber 4 (17)
1m
TC 2, TS 12
1 (100)
1 (100)
occipital 2 (9)
temporal 1 (4)
Chronic
24 (39)
7 ; 17
35.4
hypo 12 (50)
hyper 12 (50)
0 (0)
6 (50)
4 (33)
19.6
frontal 14 (58)
2m-5y
18 (75)
2 (17)
1 (9)
hyper 12 (50)
hyper 5 (21)
1 (4)
7 (58)
1 (8)
retro-bulber 6 (25)
4m-6y
TC 2, TS 16
1 (50)
1 (100)
hypo 7 (29)
1 (4)
occipital 2 (8)
parietal 1 (4)
temporal 1 (4)
Total
62
17:45
34.2
hyper 50 (81)
hyper 37 (60)
25 (40)
30 (48)
11 (18)
16.7
frontal 41 (66)
43 (69)
3 (9)
3 (9)
hypo 12 (19)
hypo 25 (40)
retro-bulber 11 (17)
TC 4, TS 39
2 (66)
2 (66)
occipital 5 (8)
temporal 4 (6)
parietal 1 (2)
Others
29
11:18
40.9
21
11
*
*
22
*
2
TC 2, TS 20
Table 1: Clinical and neuroradiological data in each type of headache.
GO term
Association gene frequency
GO frequency
molecular function unknown
189 out of 590 genes, 32.0%
189 of 1896 genes, 10.0%
hydrolase activity
82 out of 590 genes, 13.9%
82 of 861 genes, 9.5%
transferase activity
67 out of 590 genes, 11.4%
67 of 729 genes, 9.2%
structural molecule activity
43 out of 590 genes, 7.3%
43 of 345 genes, 12.5%
DNA binding
38 out of 590 genes, 6.4%
38 of 381 genes, 10.0%
RNA binding
34 out of 590 genes, 5.8%
34 of 834 genes, 4.1%
oxidoreductase activity
32 out of 590 genes, 5.4%
32 of 280 genes, 11.4%
ATPase activity
28 out of 590 genes, 4.7%
28 of 257 genes, 10.9%
enzyme regulator activity
27 out of 590 genes, 4.6%
27 of 218 genes, 12.4%
structural constituent of ribosome
22 out of 590 genes, 3.7%
22 of 224 genes, 9.8%
ion binding
19 out of 590 genes, 3.2%
19 of 176 genes, 10.8%
mRNA binding
18 out of 590 genes, 3.1%
18 of 170 genes, 10.6%
ligase activity
18 out of 590 genes, 3.1%
18 of 194 genes, 9.3%
nucleic acid binding transcription factor activity
17 out of 590 genes, 2.9%
17 of 159 genes, 10.7%
transmembrane transporter activity
17 out of 590 genes, 2.9%
17 of 313 genes, 5.4%
kinase activity
15 out of 590 genes, 2.5%
15 of 198 genes, 7.6%
protein binding transcription factor activity
15 out of 590 genes, 2.5%
15 of 126 genes, 11.9%
lipid binding
12 out of 590 genes, 2.0%
12 of 95 genes, 12.6%
cytoskeletal protein binding
11 out of 590 genes, 1.9%
11 of 66 genes, 16.7%
transcription factor binding
11 out of 590 genes, 1.9%
11 of 74 genes, 14.9%
peptidase activity
10 out of 590 genes, 1.7%
10 of 136 genes, 7.4%
nucleotidyltransferase activity
10 out of 590 genes, 1.7%
10 of 120 genes, 8.3%
unfolded protein binding
10 out of 590 genes, 1.7%
10 of 66 genes, 15.1%
phosphatase activity
10 out of 590 genes, 1.7%
10 of 94 genes, 10.6%
transferase activity, transferring glycosyl groups
9 out of 590 genes, 1.5%
9 of 96 genes, 9.4%
enzyme binding
6 out of 590 genes, 1.0%
6 of 57 genes, 10.5%
isomerase activity
6 out of 590 genes, 1.0%
6 of 58 genes, 10.3%
lyase activity
6 out of 590 genes, 1.0%
6 of 88 genes, 6.8%
GTPase activity
6 out of 590 genes, 1.0%
6 of 59 genes, 10.2%
protein transporter activity
5 out of 590 genes, 0.8%
5 of 53 genes, 9.4%
methyltransferase activity
5 out of 590 genes, 0.8%
5 of 92 genes, 5.4%
chromatin binding
5 out of 590 genes, 0.8%
5 of 101 genes, 5.0%
translation factor activity, nucleic acid binding
4 out of 590 genes, 0.7%
4 of 42 genes, 9.5%
hydrolase activity, acting on glycosyl bonds
4 out of 590 genes, 0.7%
4 of 46 genes, 8.7%
protein binding, bridging
4 out of 590 genes, 0.7%
4 of 52 genes, 7.7%
helicase activity
3 out of 590 genes, 0.5%
3 of 87 genes, 3.4%
small conjugating protein binding
3 out of 590 genes, 0.5%
3 of 44 genes, 6.8%
rRNA binding
3 out of 590 genes, 0.5%
3 of 101 genes, 3.0%
guanyl-nucleotide exchange factor activity
3 out of 590 genes, 0.5%
3 of 42 genes, 7.1%
nuclease activity
2 out of 590 genes, 0.3%
2 of 139 genes, 1.4%
histone binding
2 out of 590 genes, 0.3%
2 of 41 genes, 2.9%
signal transducer activity
2 out of 590 genes, 0.3%
2 of 39 genes, 5.1%
triplet codon-amino acid adaptor activity
0 out of 590 genes, 0%
0 of 299 genes, 0%
RNA modification guide activity
0 out of 590 genes, 0%
0 of 71 genes, 0%
other
26 out of 590 genes, 4.4%
not_yet_annotated
2 out of 590 genes, 0.3%
Table S1: Clinical and neuroradiological data in each type of headache.
GO term
Association gene frequency
GO frequency
cytoplasm
423 out of 590 genes, 71.7%
423 of 4000 genes, 10.6%
nucleus
202 out of 590 genes, 34.2%
202 of 2158 genes, 9.4%
membrane
161 out of 590 genes, 27.3%
161 of 1600 genes, 10.1%
mitochondrion
129 out of 590 genes, 21.9%
129 of 1153 genes, 11.2%
endomembrane system
81 out of 590 genes, 13.7%
81 of 774 genes, 10.5%
endoplasmic reticulum
50 out of 590 genes, 8.5%
50 of 408 genes, 12.3%
cellular component unknown
47 out of 590 genes, 8%
47 of 704 genes, 6.7%
ribosome
42 out of 590 genes, 7.1%
42 of 349 genes, 12.0%
plasma membrane
42 out of 590 genes, 7.1%
42 of 379 genes, 11.1%
mitochondrial envelope
40 out of 590 genes, 6.8%
40 of 353 genes, 11.3%
vacuole
30 out of 590 genes, 5.1%
30 of 271 genes, 11.1%
chromosome
28 out of 590 genes, 4.7%
28 of 362 genes, 7.7%
cytoskeleton
24 out of 590 genes, 4.1%
24 of 204 genes, 11.8%
site of polarized growth
23 out of 590 genes, 3.9%
23 of 239 genes, 9.6%
cellular bud
21 out of 590 genes, 3.6%
21 of 205 genes, 10.2%
nucleolus
21 out of 590 genes, 3.6%
21 of 329 genes, 6.4%
cell cortex
15 out of 590 genes, 2.5%
15 of 144 genes, 10.4%
cytoplasmic membrane-bounded vesicle
15 out of 590 genes, 2.5%
15 of 107 genes, 14.0%
Golgi apparatus
14 out of 590 genes, 2.4%
14 of 174 genes, 8.0%
cell wall
12 out of 590 genes, 2.0%
12 of 97 genes, 12.4%
peroxisome
8 out of 590 genes, 1.4%
8 of 69 genes, 11.6%
microtubule organizing center
6 out of 590 genes, 1.0%
6 of 71 genes, 8.5%
extracellular region
3 out of 590 genes, 0.5%
3 of 27 genes, 11.1%
other
11 out of 590 genes, 1.9%
not_yet_annotated
1 out of 590 genes, 0.2%
Table S2: Clinical and neuroradiological data in each type of headache.
Using the whole genome-wide ChIP on chip approach is able to provide a genome-wide scale view for the association map of interested protein or complex. Through this method, for the analysis of the genome-wide association patterns of SET3C, we obtained diverse results from 4 distinct biological repeats. That might reflect the general regulation of gene expression by SET3C and represented the dynamic role of SET3C as repressor or activator.
Acknowledgment
We thank Hsilin Cheng, Wen-Li Peng, Yi-Yun Chen, Yu-Ching Chen, Ai-Ru Wan, and lab members of Liou's lab for technical assistance. We also thank Institute of Molecular Biology, Academia Sinica (AS) for kindly providing an FEI Tecnai G2 EM, Dr. Chung- Shi Yang at Institute of Biomedical Engineering and Nanomedicine, NHRI for kindly providing a Hitachi H7650 EM, Dr. Yeu-Kuang Hwu at Institute of Physics, AS & National Synchrotron Radiation Research Center for kindly providing a Jeol 2100F EM to be used. This work was supported by grant number NSC-102-2311-B-400-002 and MG-103-PP-08.
References
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999; 98: 285-294.
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structures of the nucleosome core particle at 2.8 A resolution. Nature. 1997; 389: 251-260.
- Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005; 436: 138-141.
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001; 293: 1074-1080.
- Moazed D. Common themes in mechanisms of gene silencing. Mol Cell. 2001; 8: 489-498.
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002; 108: 489-500.
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, et al. Histone deimination antagonizes arginine methylation. Cell. 2004; 118: 545-553.
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995; 80: 583-592.
- Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006; 126: 905-916.
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001; 15: 2991-3004.
- Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R, et al. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000; 14: 1048-1057.
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000; 19: 4342-4350.
- Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci U S A. 2004; 101: 7398-7403.
- Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003; 116: 2117-2124.
- Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001; 13: 263-273.
- Cohen TJ, Mallory MJ, Strich R, Yao TP. Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot Cell. 2008; 7: 1191-1199.
- Mou Z, Kenny AE, Curcio MJ. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics. 2006; 172: 2157-2167.
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009; 137: 259-272.
- Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007; 282: 2450-2455.
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010; 39: 234-246.
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002; 298: 1412-1414.
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012; 150: 1158-1169.
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011; 42: 536-549.
- #Van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011; 475: 114-117.
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009; 457: 1033-1037.
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell. 2007; 28: 1015-1028.
- Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005; 121: 515-527.
- Chen FJ, Lee KW, Lai CC, Lee SP, Shen HH, Tsai SP, et al. Structure of native oligomeric Sprouty2 by electron microscopy and its property of electroconductivity. Biochem Biophys Res Commun. 2013; 439: 351-356.
- Tung SY, Hong JY, Walz T, Moazed D, Liou GG. Chromatin affinity-precipitation using a small metabolic molecule: its application to analysis of O-acetyl-ADP-ribose. Cell Mol Life Sci. 2012; 69: 641-650.
- Tung SY, Lee KW, Hong JY, Lee SP, Shen HH, Liou GG, et al. Changes in the genome-wide localization pattern of Sir3 in Saccharomyces cerevisiae during different growth stages. Comput Struct Biotechnol J. 2013; 7: e201304001.