
Research Article
Austin J Comput Biol Bioinform. 2015;2(1): 1011.
A Comparative Phylogenetic Evaluation of Chloroplast ITS Sequences to Analyze the Bioactivity in Medicinal Plants: A Case Study of Clerodendrum Plant Genus (Lamiaceae)
Melapu VK1, Joginipelli S1, Naidu BVA2 and Darsey J3*
1Department of Bioinformatics, University of Arkansas at Little Rock, USA
2Department of Botany, Andhra University, India
3Department of Chemistry, University of Arkansas at Little Rock, USA
*Corresponding author: Darsey J, Department of Chemistry, University of Arkansas at Little Rock, 2801 S. University Ave, Little Rock, AR
Received: March 17, 2015; Accepted: April 01, 2015; Published: April 08, 2015
Abstract
According to World Health Organization (WHO) approximately 47% of drugs used today to cure many diseases were derived from natural products such as plants. Although many approved and clinical-trial drugs have been derived from natural products, the last 20 years have shifted emphasis from natural products to less expensive synthetic products. Due to the paucity of traditional medicinal knowledge and lack of ethnobotanical information about most of the medicinal plants, the emphasis has shifted to synthetic products. Multidisciplinary research with phylogeny to incorporate ethnobotanical knowledge as well as traditional medicinal knowledge will facilitate researchers to identify new drug molecules. In this paper, we use phylogeny inferred from ITS sequence of chloroplast DNA of thirty eight different species of Clerodendrum plant to predict chemical diversity and potential medicinal activity of plants from Lamiaceae family. Phylogenetic signal in medicinal properties in plants is used to identify nodes on phylogeny with high bioscreeing potential. Phytosterol diversity and inhibition of angiotensin receptors by these compounds is significantly correlated with phylogeny. We produced a phylogenetic hypothesis for medicinally important plants of Laminaceae family based on the maximum likelihood, Neighborjoining, minimum evolution, UPGMA, and maximum parsimony methods. The genealogical trees generated with each method proved phylogenetic hypothesis by densely clustering plants with similar medicinal uses and plants belonging to same distribution. Different species of Clerodendrum plant genus were studied and their medicinal uses were evaluated in the light of phylogenetic relationships.
Keywords: WHO; Internal transcribed spacer; Phylogeny; Ethnobotanical; Clerodendrum; Lamiaceae; Bioscreeing, Phytosterol; Angiotensin receptors; Maximum likelihood; Neighbor-Joining; Minimum evolution; UPGMA; Maximum Parsimony
Abbreviations
ITS: Internal Transcribed Spacer; ME: Minimum Evolution; MEGA: Molecular Evolutionary Genetics Analysis; NCBI: National Center for Biotechnology Information; NJ: Neighbor-Joining; OLS: Ordinary Least Sqaures; OUT: Operational Taxonomy Units), UPGMA: Unweighted Pair Group Method with Arithmetic mean; WHO: World Health Organization
Introduction
Throughout the ages, humans have relied on nature for their basic needs such as food, shelter, clothing and medicines. Due to the natural ability of plants to synthesize wide variety of compounds; they are used as source of medicines to cure ailments and prevent diseases. Countries with ancient civilization such as India, China, Egypt, etc used plants as remedies for various conditions and ailments. According to World Health Organization (WHO) approximately 25% of drugs used today to cure many diseases were derived from plants and plant substances [1]. Researchers across the globe from different regions of the world have worked on medicinal plants to identify and discover new molecules and to make advances in production of drugs derived from plant products. Ethnobotanical knowledge about plants and their species is essential for further investigations to discover new molecules from plants. Compiling a comprehensive list of medicinal plants and their species across different regions of the world will preserve our knowledge about the medicinal plants and will facilitate researchers to identify new molecules [2, 3].The field of bio-prospecting and bioscreening guided by the ethnomedicinal knowledge from traditional medicine, led to the discovery of several plant derived pharmaceuticals such as Taxol, Chloroquine, Crystodigin, Galanthamine, and Nitisinone etc [4]. Chemotherapy drug Taxol used in current treatment for breast cancer, ovarian cancer and lung cancer is derived from chemical obtained from Taxus brevifolia plant [5]. Ancient Egyptians used cinchona bark or bark from willow tree to treat and cure malarial fevers, active chemical compound quinine was extracted and is still used in treatment of malarial fevers. Crystodigin and Lanoxin drugs used in treatment of heart conditions such atrial fibrillations and congestive heart failure, are derived from digitoxin obtained from Digitalis purpurea (Floxglove) plant [6]. Although many approved and clinical-trial drugs have been derived from natural products, the last 20 years have shifted emphasis from natural products to less expensive synthetic products. Therefore, improvements in bioprospecting on the basis of phylogenetic methods have very recently been given considerable attention by mostly Chinese medicinal plant chemists, British biologists, and some ethnobotanists. One approach to increasing drug discovery via natural products is to use phylogenetic methods (“genealogical focusing”) to locate lineages on the “tree of life” that are most likely to yield useful drug-like scaffolds [7].
Clerodendrum plant genus of about 400 to 500 species, belonging to Lamiaceae family is a subtropical and tropical plant mostly distributed in Africa, Asia and Pacific Oceania [8]. The genus comprising of about 400 to 500 species shows high degree morphological variation and has many medicinal uses. The medicinal properties are not randomly distributed in plants, some group of plants show high medicinal properties than the other group. This suggests that there is a phylogenetic pattern of medicinal properties across the world and is revealed with phyglogenetic analysis [9]. Here in this study we have performed phylogenetic analysis on thirty eight different species of Clerodendrum plant to understand and evaluate the cross cultural patterns, medicinal properties and chemical diversity. We have inferred the phylogeny using ITS (Internal Transcribed Spacer) regions of chloroplast DNA.
Materials and Method
Collection of medicinal plant species of clerodendrum
Some species of Clerodendrum plant such as Clerodendrum infortunatum, Clerodendrum serratum, Clerodendrum viscosum, etc. were collected from (Western Ghats of India, Coimbatore, Hyderabad, India) southern region of India, with the help of Dr. B.V.A Rama Rao Naidu, Botanists and Taxonomist. A voluntary survey about the medicinal plants was conducted to know the medicinal properties of the plants from medical practitioners of the study region. Some species of Clerodendrum were retrieved from extensive literature search including ethnobotanical databases, published research articles. We collected information on the medicinal applications of Clerodendrum throughout the range of the genus. Table 1 shows the thirty eight different species of Clerodendrum genus, its medicinal uses, and the geographical distribution of the species in the region (Table 1).
Scientific Name of the plant
Medical uses
Region
Clerodendrum buchanani
Laxative, gastrointestinal disorders
Malaysia, Hawaii
Clerodendrum bipindense
Antiinflammatory, Analgesic
West Tropical Africa
Clerodendrum bungei
Antimicrobial, Antiinflammatory
India, China
Clerodendrum canescens
Antihypertensive
China, Taiwan
Clerodendrum capitatum
Antihypertensive, Antidiabetic,
Erectile dysfunction
Africa
Clerodendrum cephalathum
sub species mashariki
Eye problems
Africa
Clerodendrum cephalanthum
sub species montanum
Eye problems
Africa
Clerodendrum chinense
Antihypertensive, Antimicrobial, Antiinflammatory, Analgesic
China
Clerodendrum colebrookianum
Antihypertensive, Cough and Dysentry
India, Myanmar, Bangladesh, Malaysia, and Nepal
Clerodendrum cyrtophyllum
Antipyretic, Diuretic
China
Clerodendrum floribundum
Laxative, gastric disorders
Australia, New Guinea
Clerodendrum fortunatum
Analgesic
India, China
Clerodendrum fragrans
Analgesic
India
Clerodendrum glabrum
Antihypertensive
Africa
Clerodendrum hildebrandtii
Skin diseases
Africa
Clerodendrum indicum
Not-known
India, china
Clerodendrum infortunatum
Antimicrobial, Antifungal
India, Sri Lanka
Clerodendrum japonicum
Antimicrobial, Antihypertensive
Japan, India, China
Clerodendrum johnstonii
Not known
Africa
Clerodendrum lindleyi
Not known
China
Clerodendrum mandarinorum
Antihypertensive
China
Clerodendrum minahassae
Gastric disorders
China, India
Clerodendrum paniculatum
Not known
India, Sri Lanka, Bangladesh
Clerodendrum phlomidis
Antiinflammatory
China, India
Clerodendrum poggei
Ornamental garden plant
Australia
Clerodendrum polycephalum
Analgesic, Antiparalytic
Africa
Clerodendrum quadriloculare
Ornamental garden plant
New-Guinea, Philippines
Clerodendrum rotundifolium
Analgesic
Africa
Clerodendrum schweinfurthii
Analgesic
Africa
Scientific Name of the plant
Medical uses
Region
Clerodendrum buchanani
Laxative, gastrointestinal disorders
Malaysia, Hawaii
Clerodendrum bipindense
Antiinflammatory, Analgesic
West Tropical Africa
Clerodendrum bungei
Antimicrobial, Antiinflammatory
India, China
Clerodendrum canescens
Antihypertensive
China, Taiwan
Clerodendrum capitatum
Antihypertensive, Antidiabetic,
Erectile dysfunction
Africa
Clerodendrum cephalathum
sub species mashariki
Eye problems
Africa
Clerodendrum cephalanthum
sub species montanum
Eye problems
Africa
Clerodendrum chinense
Antihypertensive, Antimicrobial, Antiinflammatory, Analgesic
China
Clerodendrum colebrookianum
Antihypertensive, Cough and Dysentry
India, Myanmar, Bangladesh, Malaysia, and Nepal
Clerodendrum cyrtophyllum
Antipyretic, Diuretic
China
Clerodendrum floribundum
Laxative, gastric disorders
Australia, New Guinea
Clerodendrum fortunatum
Analgesic
India, China
Clerodendrum fragrans
Analgesic
India
Clerodendrum glabrum
Antihypertensive
Africa
Clerodendrum hildebrandtii
Skin diseases
Africa
Clerodendrum indicum
Not-known
India, china
Clerodendrum infortunatum
Antimicrobial, Antifungal
India, Sri Lanka
Clerodendrum japonicum
Antimicrobial, Antihypertensive
Japan, India, China
Clerodendrum johnstonii
Not known
Africa
Clerodendrum lindleyi
Not known
China
Clerodendrum mandarinorum
Antihypertensive
China
Clerodendrum minahassae
Gastric disorders
China, India
Clerodendrum paniculatum
Not known
India, Sri Lanka, Bangladesh
Clerodendrum phlomidis
Antiinflammatory
China, India
Clerodendrum poggei
Ornamental garden plant
Australia
Clerodendrum polycephalum
Analgesic, Antiparalytic
Africa
Clerodendrum quadriloculare
Ornamental garden plant
New-Guinea, Philippines
Clerodendrum rotundifolium
Analgesic
Africa
Clerodendrum schweinfurthii
Analgesic
Africa
Table 1: Multiple sequence alignment of Clerodendrum species with Clustax 2.1.
Collection of sequence data
ITS (Internal Transcribed Spacer) regions from chloroplasts of thirty eight different species were collected from NCBI database (National Center for Biotechnology Information) [10]. Due to different rates of evolution, nuclear ribosomal internal transcribed spacer regions have become the routine marker in evolutionary studies at different taxonomic levels in plants. Internal Transcribed Spacers are within the ribosomal transcript that are excised and degraded during maturation; their sequences generally show more variation than the ribosomal sequence, making them popular for phylogenetic analysis and or identification of species and strains. The nucleotide sequences in FASTA format were used to construct the phylogenetic trees. The nucleotide sequences were used to do the multiple sequence alignment using clustax 2.1 [11]. The multiple sequence alignment from 50 to 270 nucleotide positions is displayed on the Clustax window is shown in (figure 1), the alignment is color coded (Figure 1).
Figure 1: Multiple sequence alignment of Clerodendrum species with Clustax 2.1.
Phylogenetic analysis
The alignment file generated from multiple sequence alignment is used to generate the phylogentic tree for different species; phylogeny is performed with MEGA (Molecular Evolutionary Genetics Analysis) [12]. The phylogenetic trees were generated using maximum likelihood method, Neighbor-Joining method (NJ), Minimum Evolution (ME) method, UPGMA (Unweighted Pair Group Method with Arithmetic mean) method, and maximum parsimony method. One letter genetic code for the commonly used genetic tables is used in the display, figure 2 shows the maximum parsimonious sites, the maximum parsimonious sites were highlighted with yellow color. The most common sites, most parsimonious sites, most variable sites and most singleton sites can be highlighted using the options in MEGA (Figure 2).
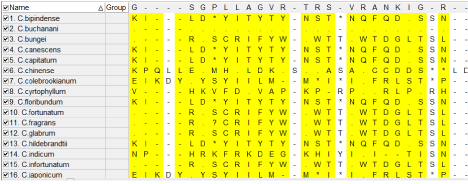
Figure 2: Molecular data explore showing most parsimonious sites highlighted in yellow for sixteen species of Clerodendrum plant.
Results and Discussion
Maximum likelihood is general statistical method for estimating the unknown parameters of a probability model. A parameter is some descriptor of the model, for normal distribution model it is the mean and variance. In phylgenetics there are many parameters such as rates, differential transformation cost, and tree itself. Likelihood is a quantity proportional to the probability of observing the data. It evaluates a hypothesis about evolutionary history in terms of the probability that the proposed model and the hypothesized history would give rise to the observed data set. The supposition is that a history with a higher probability of reaching the observed state is preferred to a history with a lower probability. The method searches for the tree with the highest probability or likelihood [12]. Here the maximum likelihood phylogenetic tree generated is shown in figure 3. Plants with similar medicinal uses for related health conditions in relatively independent regions of the world will converge into one single branch when compared to plants with varying medicinal uses, and different pharmacological actions. These types of medicinal uses that are most likely reflect DNA phylogeny and correlate with other shared derived characters. It is expected that such medicinal uses are more likely to clump on phylogeny. It is expected that plants with similar medicinal uses will tend to more densely cluster around specific lineages rather than scatter randomly throughout the tree. Therefore, we can say plants with general disorder category of medicinal uses (e.g. GIT conditions) in traditional medicine will tend to cluster together in broader lineages rather than being scattered more randomly.
Figure 3: Phylogenetic tree generated with maximum likelihood method for
thirty eight species of Clerodendrum plant.
The tree generated with maximum likelihood in broader view displayed two different lineages; six different species of Clerodendrum genus; C. colebrookianum, C. japonicum, C. splendens, C. trichotomum, C. yunnanense, and C. chinense branches out into one lineage and the other thirty two species branched out into one broader lineage as shown in figure 3. From table 1 we can say the six different species of Clerodendrum genus, C. colebrookianum, C. japonicum, C. splendens, C. trichotomum, C. yunnanense, and C. chinense have same medicinal use i.e. antihypertensive’s to relieve the symptoms of hypertension. Tree generated with maximum likelihood differentiated plants with antihypertensive medicinal use from the rest of the plants with different medicinal uses. These plant species have more concentrations of phytosterol compounds such as campsterol, Beta-sitosterol, cholestanol, clerosterol used as antihypertensive due to their ability to inhibit angiotensin type II receptors in hypertension clustered into single branch in phylogeny indicating the chemical similarity (Figure 3).
Neighbor-Joining (NJ) method is a method of reconstructing phylogenetic trees, and computing the lengths of the branches of the tree. In each stage, the two nearest nodes of the tree are chosen and defined as neighbors in the tree. Neighbors are a pair of OTU’s (Operational Taxonomic Units), and are used to construct the trees, two nodes with smallest distance will be considered for starting the generation of the tree and the tree will be expanded until all the nodes are incorporated [12]. Neighbor Joining method seeks to build a tree which minimizes the sum of all edge lengths, i.e., it adopts the Minimum Evolution (ME) criterion. For small number of taxa, NJ solutions are likely to be identical to the optimal ME tree [13]. As the number of taxa or number species of Clerodendrum plant were less, the tree generated with NJ method is identical to the tree generated with ME method. The trees generated from NJ and ME method are shown in figure 4 and figure 5 with branch lengths and nodal ids. In both NJ and ME method, seven different species of Clerodendrum; C. cryptophyllum, C. phlomidis, C. schweinfurthii, C. minahassae, C. mandarinorum clustered into one single lineage. The distances and branch lengths of both NJ and ME methods showed 98% of correlation, with similar branch lengths and nodal lenghts. All the seven different species were distributed in India and China. C. buchanani and C. minahassae with related medicinal uses, i.e. Gastric disorders clustered in to on sub branch and C. Phlomidis and C. schweinfurthii with related medicinal uses, i.e. anti-inflammatory uses clustered into one sub branch (Figure 4).
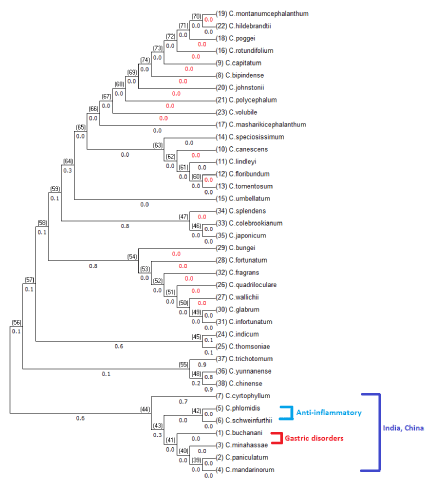
Figure 1: Phylogenetic tree generated with Neighbor-Joining method for
thirty eight different species of Clerodendrum plant.
Minimum evolution method of generation of phylogenetic tree inference is based in the assumption that the tree with the smallest sum of branch length estimates is most likely to be the best tree. The branch lengths are estimated by Ordinary Least Squares method (OLS), from the distance matrices. This algorithm uses NJ tree to start with nodes and then expands with OLS using mathematical proofs [14], (Figure 5).
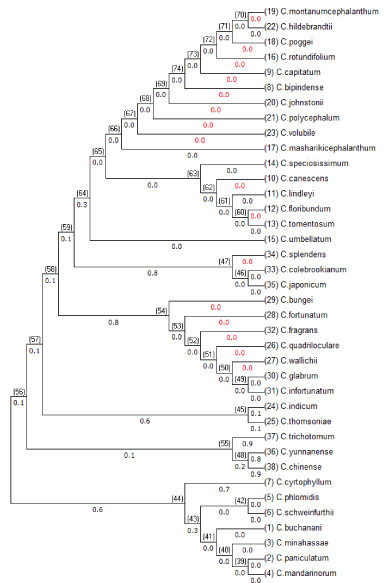
Figure 5: Phylogenetic tree generated with minimum evolution method for
thirty eight species of Clerodendrum plant.
UPGMA (Unweighted Pair Group Method with Arithmetic Mean) is a simple agglomerative bottom up method of hierarchical clustering. UPGMA assumes a constant rate of evolution and is not well regarded method of inferring relationships unless the assumption is tested and justified. Here at each step the nearest two clusters are combined into a higher level cluster [15]. The tree generated from UPGMA method differentiated the lineages according to their habitat and distribution. Plant species belonging to one habitat of distribution in the world clustered into relatively single branch, as shown in the figure 6 (Figure 6).
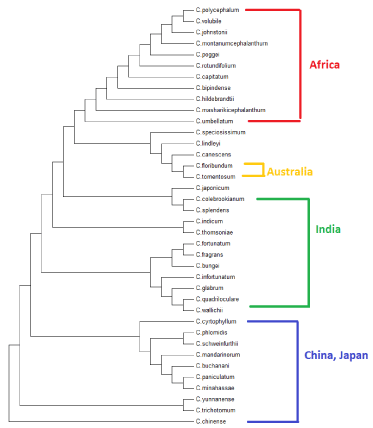
Figure 6: Phylogenetic tree generated with UPGMA method for thirty eight
species of Clerodendrum plant.
Maximum parsimony is a non parametrical statistical method, and the trees that have the least evolutionary change to explain the observed data will be used, hence they are maximally parsimonious. This is a character based tree estimation method and uses a matrix of discrete phylogenetic characters to infer the optimal trees for a set of taxa or species [16]. The results generated with maximum parsimony are shown in figure 7. Maximum parsimony method will result in more than one parsimonious results, so the consensus tree which includes the results from all the parsimonious trees is generated with boot strapping method. The consensus tree generated is shown in figure 8. Maximum parsimony tree shown in figure 7 and consensus tree shown in figure 8 densely clustered the antihypertensive species of Clerodendrum supporting the results generated from maximum likelihood method (Figure 7).
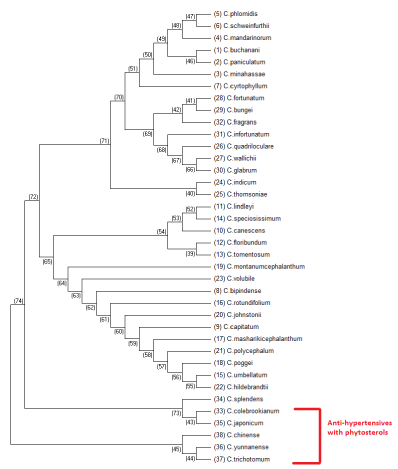
Figure 7: Phylogenetic tree generated with maximum parsimony method for
thirty eight different species of Clerodendrum plant.
For the generation of consensus tree 3 most parsimonious trees were used and tree #1 the best tree (length = 1207) is shown in figure 8. The consistency index is 0.587407 (0.586035), the retention index is 0.852444 (0.852444), and the composite index is 0.500732 (0.499562) for all sites and parsimony-informative sites (in parentheses). The MP tree was obtained using the Sub tree-Pruning-Regrafting (SPR) algorithm [17] with search level 0 in which the initial trees were obtained by the random addition of sequences (10 replicates). The analysis involved 38 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 307 positions in the final dataset (Figure 8).
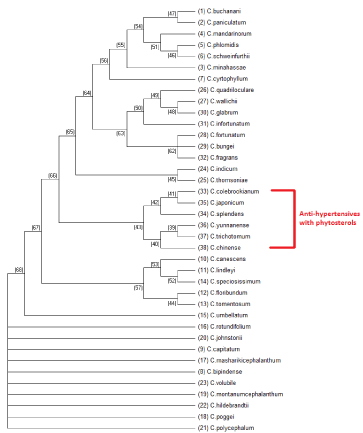
Figure 8: Phylogenetic consensus tree generated with boot strapping the
maximum parsimony tree for thirty eight species of Clerodendrum plant
genus.
Conclusion
The bioprospecting and the proposed hypothesis about the possible clustering of the phylogenies across the species of Clerodendrum were done based on the ethnobotanical inputs collected directly from the study region, and from the voluntary survey of medical practitioners of the study region. The results of the phylogenetic clustering validated the hypothesis. The incorporation of some species of Clerodendrum, which are native to the tribal population into the phylogenetic analysis has provided the platform for exploring the medicinal properties of some more Clerodendrum species never studied before. This study is centered on the possible identification of some other Clerodendrum species with wide range of different medicinal properties in the northeastern ghats of southern India. This study will guide us into identification of new species of Clerodendrum plant with some medicinal properties and have possible implications on enlarging Lamiaceae family with addition of some species.
We have shown a significant correlation between phylogeny, chemical diversity and medicinal activity or biological activity of the Clerodendrum plant genus belonging to Lamiaceae family. This research is multidisciplinary, which incorporates ethnobotanical, biogeographical, bioactivity, and phylogenetic information to provide new perspectives in bioactivity of medicinal plants based on the traditional usage, bio-geographical distribution, and phylogenetic conservatism. Similar medicinal uses for related health conditions in relatively independent regions of the world clustered with a quantitatively high degree of fit to the same plant lineages on the phylogenetic trees. Plants belonging to same region in the distribution by habitat clustered more densely than the other plants belonging to different geographical distribution. Clerodendrum plant species with antihypertensive usage clustered densely when compared to other medicinal uses, and the species of plants belonging to Africa clustered together, and where as plants distributed across Asia clustered together and denser clusters were seen for plants belonging to India, China and Japan. The analysis of the phylogeny of the Clerodendrum species provided us the necessary insight into the bioactive compounds, and provides the basis for the discovery of new drug leads. The distances mapped using NJ method and ME method showed similar results with approximately 98% of correlation. The distances and branch lengths mapped using UPGMA showed dense clusters belonging to same bio geographical distribution. The maximum parsimony and maximum likelihood methods mapped dense clusters of plant species with same medicinal activity i.e. antihypertensive, which can inhibit the angiotensin type II receptors.
Phylogeny can be used to interpret and predict the chemical activity and bioactivity of the plants and plant related products and can further lead to lead drug discovery. The very recent use of plant phylogeny in conjunction with traditional medical knowledge may help us to locate new drugs from traditional medicinal plants and can further lead to discovery of novel drug molecules for the treatment of many diseases. Phylogeny or genealogical focusing is the best approach to increase the drug discovery via natural products to locate lineages that are most likely to yield useful drug like scaffolds. Bio prospecting combined with phylogeny, ethnobotanical knowledge and traditional medicinal knowledge will aid the process of discovery of drugs from natural products such as plants.
Acknowledgement
This project was supported by grants from the National Center for Research Resources (5P20RR016460-11) and the National Institute of General Medical Sciences (8 P20 GM103429-11) from the National Institutes of Health.
References
- "Traditional Medicine: Definitions and plant derived drugs". World Health Organization. 2008-12-01.
- Clarkson C, Maharaj VJ, Crouch NR, Grace OM, Pillay P, Matsabisa MG, Bhagwandin N. In vitro antiplasmodial activity of medicinal plants native to or naturalised in South Africa. J Ethnopharmacol. 2004; 92: 177-191.
- Fabricant DS, Farnsworth NR. The value of plants used in traditional medicine for drug discovery. Environ Health Perspect. 2001; 109 Suppl 1: 69-75.
- Wright CI, Van-Buren L, Kroner CI, Koning MM. Herbal medicines as diuretics: a review of the scientific evidence. J Ethnopharmacol. 2007; 114: 1-31.
- Topliss JG, CLARK AM, Ernst E, Hufford CD, Johnston GA. R, Weimann BJ, et al. Natural and synthetic substances realated to human health. International Union of Pure and Applied Cheimstry 2002. 74: 1957-1985.
- Mulabagal Vanisree, Chen-Yue Lee, Shu-Fung Lo, Satish Manohar Nalawade, Chien Yih Lin, Hsin-Sheng Tsay. Studies on the production of some important secondary metabolites from medicinal plants by plant tissue cultures. Botanical Bulletin of Academia Sinica. 2004. 45: 1-22.
- Zhu F, Qin C, Tao L, Liu X, Shi Z, Ma X, Jia J. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Natl Acad Sci U S A. 2011; 108: 12943-12948.
- Stenzel E, Heni J, Rimpler H, Vogellehner D. Phenetic relationships in Clerodendrum (Verbenaceae) and some phylogenetic considerations. Plant Systematics and Evolution 1988; 159: 257–271.
- Bennett BC, Husby CE. Patterns of medicinal plant use: an examination of the Ecuadorian Shuar medicinal flora using contingency table and binomial analyses. J Ethnopharmacol. 2008; 116: 422-430.
- NCBI, (National Center for Biotechnology Information).
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23: 2947-2948.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30: 2725-2729.
- Saitou, N. Imanishi T. Relative efficiencies of the Fitch-Margoliash, maximum-parsimony, maximum-likelihood, minimum-evolution, and neighbor-joining methods of phylogenetic tree construction in obtaining the correct tree. Mol. Biol. Evol. 1989; 6: 514–525.
- BULMER, M. Use of the method of generalized least squares in reconstructing phylogenies from sequence data. Mol. Biol. Evol. 1991; 8:868-883.
- Sokal R and Michener C. "A statistical method for evaluating systematic relationships". University of Kansas Science Bulletin.1958; 28: 1409-1438.
- J. Felsenstein."Cases in which parsimony and compatibility methods will be positively misleading". Systematic Zoology.1978; 27: 401–410.
- Nei M. and Kumar S. Molecular Evolution and Phylogenetics. Oxford University Press, New York.2000.