
Research Article
J Dent & Oral Disord. 2021; 7(1): 1154.
RASSF1A and KRAS Expression in Oral Leukoplakia with Dysplasia and in Squamous Cell Carcinoma
Costa V1, El-Achkar VNR1, Ribeiro MP1, Tristao SS1, Kowalski LP2, Pinto CAL3, Carvalho YR1, da Cruz-Perez DE4 and Kaminagakura E1*
1Department of Biosciences and Oral Diagnosis, Sao Paulo State University (Unesp), Brazil
2Department of Head and Neck Surgery, University of Sao Paulo Medical School, Brazil
3Department of Anatomic Pathology, AC Camargo Cancer Center, Brazil
4Department of Clinical and Preventive Dentistry, Pernambuco Federal University (UFPE), Brazil
*Corresponding author: Kaminagakura E, Department of Biosciences and Oral Diagnosis, Sao Paulo State University (Unesp), Brazil
Received: January 21, 2021; Accepted: February 17, 2021; Published: February 24, 2021
Abstract
The aim of this study is to investigate the role of RASSF1A and KRAS protein immunoexpression in Oral Leukoplakia with Epithelial Dysplasia (OLD) and in Oral Squamous Cell Carcinoma (OSCC). Immunohistochemical staining for RASSF1A and KRAS was performed and a semiquantitative analysis was applied to samples of the Control Group (CG, n=20), OLD (n=39), and OSCC (n=100). No significant difference was observed between RASSF1A immunoexpression and OLP and OSCC groups (p>0.05). KRAS expression was higher in OSCC than in OLP and CG (p<0.05). No association was observed between RASSF1A or KRAS expression and alcohol/tobacco use or clinicopathological features (p>0.05) in the OSCC group. Also, patients with OSCC who presented KRAS overexpression had a worse disease-free survival rate (p=0.04). RASSF1A expression was similar in OLD and OSCC groups, suggesting that it plays a critical role in the early stage of OSCC. KRAS expression was higher in OSCC when compared with normal and dysplastic tissues, showing that KRAS expression increases with malignant progression.
Keywords: Oral leukoplakia; Oral cancer; Oral squamous cell carcinoma; Oncoprotein
Introduction
Oral leukoplakia is recognized as a white plaque of questionable risk, diagnosed excluding other known disorders or diseases that carry no increased risk for cancer [1]. It is considered as the most common potentially malignant oral disorder encountered in clinical practice [2,3]. Its malignant transformation is reported in about 1% of cases, resulting in 20 per 100,000 new cases of Oral Squamous Cell Carcinoma (OSCC) per year [2].
Squamous Cell Carcinoma (SCC) is the most common malignant neoplasm of the oral cavity. Alcohol abuse and tobacco use are the major risk factors for oral cancer [4]. They increase the Reactive Oxygen Species (ROS) causing oxidant/antioxidant imbalance. This is accompanied by lipid peroxidation, oxidative DNA damage, damage to macro- and micro-molecules of cells, and disturbances of antioxidant defense, which can initiate the malignant process [5].
Mutant KRAS (Kirsten Murine Sarcoma Virus) elevates intracellular Reactive Oxygen Species (ROS) levels and leads to oxidative DNA damage. Ras Association Domain Family 1 Isoform A (RASSF1A) is known to play a role as a Ras effector. The suppressive effect of RASSF1A on ROS production is triggered by activated K-RAS. RASSF1A attenuates KRAS-triggered oxidative DNA damage and chromosomal damage [6].
Despite advances in diagnostic methods, the malignant transformation predictor of oral leukoplakia into OSCC is still based on conventional histopathological examination [3]. In recent years, the interest in genes that predispose to the carcinogenesis of oral leukoplakia has grown. Therefore, the aims of this study are to investigate the pattern of RASSF1A and KRAS immunoexpression in Oral Leukoplakia with Epithelial Dysplasia (OLD) and compare to OSCC and to epithelial without dysplasia (control group/CG), and to associate the results with clinicopathological features.
Materials and Methods
The Research Ethical Committee of the University of Sao Paulo State approved this study (CAAE 34246314.9.1001.0077). Previously, calibrated examiners performed all the following analyzes independently and the Kappa test was used to determine agreement between them (VC, MPR and EK).
A retrospective analysis from 1990 to 2007 was performed and 100 samples with diagnosis of OSCC were obtained from the files of the Department of Pathology, AC. Camargo Cancer Center, Sao Paulo, Brazil. During a period from 1992 to 2016, 39 samples with diagnosis of OLD and 20 samples of epithelial tissue from fibroma without epithelial dysplasia or inflammation (control group/CG) were obtained from the files of the Department of Biosciences and Oral Diagnosis, São Paulo State University, São José dos Campos and from Department of Clinical and Preventive Dentistry, Pernambuco Federal University, Brazil. Clinicopathological features, variables and follow-ups were collected from patients’ medical history charts. Cases of OSCC were staged according to the AJCC cancer staging manual [7]. Histopathological diagnoses and histological grades were reviewed and were classified according to El-Naggar et al., 2017 [4].
Tissue Microarray (TMA)
For OSCC samples, TMA was built following methods similar to those reported earlier by Kaminagakura et al., 2011 [8].
Immuno HistoChemistry (IHC)
The methods used herein are similar to those reported earlier by Costa et al., 2019 [9]. The sections were incubated with primary antibodies against RASSF1A (1:200, clone NBP1-89411, Novus Biologicals, CO, EUA) and KRAS (1:100, clone ab55391, Abcam, MA, USA) at 4°C overnight.
A semiquantitative analysis using an Olympus CX31 light microscope (Olympus, New York, Ny, USA) with a 10x/0.25 objective was performed. For IHC analysis, less than 30% of the cytoplasmic immunostained positive cells were considered underexpressed, and ≥30% were considered overexpressed according to the modified criteria proposed by Cao et al., 2013 [10] and Elsabah et al., 2013 [11].
Statistical analysis
Statistical analyses were performed using the SPSS program, version 23.0 (SSPInk, IL, USA). The association between qualitative variables was evaluated by X2 test or Fisher’s exact test, as appropriate. For OSCC, the Kaplan-Meier estimator of the survival function was considered for survival analysis. Only results with p<0.05 were considered statistically significant.
Results
The clinicopathological data and immunoexpression status are summarized in (Table 1).
Characteristics
CG
OLD
OSCC
n (%)
Age, years
Mean
Range44.2
14-7151.2
19-8050
20-80Sex
Male
9 (45.0%)
21 (53.9%)
74 (74.0%)
Female
11 (55.0%)
18 (46.1%)
26 (26.0%)
Race
White
Other19 (95.0%)
1 (5.0%)29 (74.4%)
10 (25.6%)82 (82.0%)
18 (18.0%)Location
Tongue
Lip mucosa
Floor of the mouth
Retromolar area
Buccal mucosa
Palate
Gengiva
Alveolar ridge
Other6 (30.0%)
4 (20.0%)
*
*
10 (50.0%)
*
*
*
05 (12.8%)
1 (2.6%)
7 (18%)
*
9 (23.0%)
6 (15.4%)
4 (10.2%)
5 (12.8%)
2 (5.2%)47 (47.0%)
*
26 (26.0%)
15 (15.0%)
2 (2.0%)
*
*
*
10 (10.0%)Tobacco use
Tobacco exposure
No tobacco exposure
Not reported*
*
**
*
*77 (77.0%)
16 (16.0%)
7 (7.0%)Alcohol use
Alcohol exposure
No alcohol exposure
Not reported*
*
**
*
*57 (57.0%)
36 (36.0%)
7 (7.0%)T classification
T1 - T2
T3 - T4*
**
*39 (39.0%)
61 (61.0%)N classification
N0
N+*
**
*43 (43.0%)
57 (57.0%)M classification
M0
M+*
**
*98 (98.0%)
2 (2.0%)Histological grade
Mild/Well differentiated
Moderate/Moderately differentiated
Severe/Poorly differentiated
Not reported*
*
*
*24 (61.5%)
9 (23.1%)
6 (15.4%)
*58 (58.0%)
26 (26.0%)
11 (11.0%)
5 (5.0%)RASSF1A
Overexpression
Underexpression
Lost samples0 (0.0%)
18 (100%)
226 (68.5%)
12 (31.5%)
164 (67.4%)
31 (32.6%)
5KRAS
Overexpression
Underexpression
Lost samples4 (20.0%)
16 (80.0%)
09 (23.7%)
30 (76.3%)
060 (71.4%)
24 (28.6%)
16*Not applicable or information not reported.
Table 1: The clinicopathological data and immunoexpression status are summarized.
Control group (CG)
The control group consisted of irritation fibroma without epithelial dysplasia or inflammation in the connective tissue. The mean age was 44.2 years old (range: 14 to 71 years old), 11 (55.0%) patients were female, 10 (50.0%) cases were located mostly in the buccal mucosa, 6 (30.0%) in the tongue and 4 (20.0%) cases in lip mucosa.
In the IHC analysis, RASSF1A underexpression was observed in all cases (100%) (Figure 1A), and 2 cases were lost. KRAS overexpression was present in 4 (20.0%) cases and underexpression was observed in 16 (80.0%) cases (Figure 1B). RASSF1A expression was statistical lower in CG than in OLP and OSCC (p<0.05).
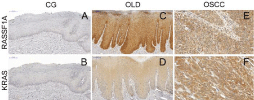
Figure 1: RASSF1A and KRAS expression in Control Group (CG), Oral
Leukoplakia with Dysplasia (OLD), and Oral Squamous Cell Carcinoma
(OSCC). A) No RASSF1A expression was observed in CG; B) KRAS
underexpression in CG; C) RASSF1A expression in OLD, showing a
positivity in both basal and suprabasal layers of the epithelial cells; D) KRAS
expression in OLD, presenting positivity in both basal and suprabasal layers;
E) RASSF1A expression in OSCC, showing cytoplasmatic overexpression
in tumor cells; F) KRAS expression exhibiting a cytoplasmatic positivity in
OSCC cells.
Oral Leukoplakia with Epithelial Dysplasia (OLD)
The mean age was 51.2 years old (range: 19 to 80 years) with male prevalence: 21 (53.9%) cases. Most patients in this group were white, with 29 (74.4%) cases, and the lesions were located mostly in the buccal mucosa with 9 (23.0%) cases, followed by floor of the mouth with 7 (18%) cases. Microscopically, 24 (61.5%) cases presented mild epithelial dysplasia, 9 (23.1%) presented moderate epithelial dysplasia, and 6 (15.4%) exhibited severe epithelial dysplasia.
In the IHC analysis, RASSF1A over and underexpression were observed in 26 (68.5%) and in 12 (31.5%) cases, respectively (Figure 1C). One sample was lost and excluded from the RASSF1A analysis. KRAS overexpression was noted in 9 (23.7%) cases and underexpression was observed in 30 (76.3%) cases (Figure 1D).
Oral Squamous Cell Carcinoma (OSCC)
The mean age was 50 years old (range: 20 to 80 years), with a male prevalence of 74 (74%) cases. Most patients in this group were white, numbering 82 (82%) cases. The lesions were located mostly in the oral tongue, with 47 (47%) cases, followed by floor of the mouth with 26 (26%) cases. In total, 77 (77%) patients were smokers and 57 (57%) were alcohol drinkers. According to the tumor size, 39 (39%) cases were T1-T2, and 61 (61%) cases were T3-T4. There was lymph node involvement (N+) in 57 (57%) cases. Only 2 (2%) patients had distant metastasis. Well-differentiated tumors were observed in 58 (58%) cases.
In the IHC analysis, RASSF1A overexpression was observed in 64 (67.4%) cases and KRAS overexpression was noted in 60 (71.4%) cases (Figure 1E&1F). In addition, RASSF1A underexpression was present in 31 (32.6%) cases and KRAS underexpression was observed in 24 (28.6%) cases. No association was observed between RASSF1A or KRAS expression and alcohol/tobacco use or other clinicopathological features (p>0.05), in this group. KRAS expression was higher in OSCC than in OLP and CG (p<0.05). OSCC patients who presented KRAS overexpression had the worst disease-free survival curve (p=0.04) (Figure 2).
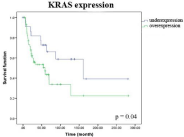
Figure 2: Disease-free survival curve of OSCC patient with KRAS
overexpression (p=0.04).
Discussion
In the current study, we have not found association between RASSF1A expression with alcohol abuse, tobacco use, OSCC location or gender. It was underexpressed in CG but overexpressed in OLD and OSCC. RASSF1A protein was expressed more in head and neck SCC than in its surgical margins, which might indicate either high risk of mortality among the examined patients or the fact that the role of RASSF1 in tumorigenesis is more complex [12] than was supposed. However, the RASSF1A protein expression in oral SCC and adjacent tissues was significantly lower than that in control tissues [13].
In vitro study, knockdown of RASSF1A gene in tongue squamous cell carcinoma cell line promoted proliferation, migration and inhibited apoptosis of cancer cells. It is suggested that the RASSF1A gene may be involved in the proliferation and metastasis of OSCC [13]. At the same time, in mice, overexpression of the RASSF-1A gene reduced tumor growth-inhibiting CyclinD1 protein expression [13]. CyclinD1 overexpression was correlated with worse diseasefree survival without its gene amplification [8]. Probably, CyclinD1 protein induction during cell cycle progression from S to G2 phase is Ras-activity dependent [14]. In this study, there is no association between RASSF1A expression with the survival rates, contradicting Sun et al., 2019 [13] who reported the high level of RASSF1A transcripts was significantly associated with recurrence-free survival using 50% and 25% cut-off thresholds.
In OSCC, RASSF1A plays a role as a Ras effector, modulating reactive oxygen species and preventing chromosomal and oxidative DNA damage induced by oncogenic KRAS [6]. The main factors risk for OSCC: tobacco and alcohol, increase Reactive Oxygen Species (ROS) formation, causing oxidant and antioxidant imbalance [5]. In mice, nicotine induced dedifferentiation of pancreatic acinar cells by activating AKT-ERK-MYC signaling; this led to inhibition of GATA6-promoter activity, loss of GATA6 protein, and subsequent loss of acinar differentiation and hyperactivation of oncogenic KRAS; favoring pancreatic cancer progression. At the same time, there was a significant increase in the number of proliferating acinar cells expressing Aldehyde Dehydrogenase (ALDH) [15]. ALDH is an enzyme responsible for metabolizing ACTH; it can cause DNA damage and favor cancer progression [16]. The ALDH1A1 overexpression in OLD has been reported, but it was absent in OSCC, suggesting an important role in the early onset of malignant transformation [9,17]. Otherwise, the ADLH2 was overexpressed in OSCC [17].
Hypermethylation of RASSF1A can be detected in both early stage and advanced nasopharyngeal carcinoma, suggesting that the RASSF1A gene promoter methylation might play an important role in the early development of its carcinogenesis. RASSF1A expression could inhibit tumorigenicity through induction of cell cycle arrest in the G0/G1 phase, and mediated apoptosis in a Ras-dependent manner. RASSF1A inactivation is essential for nasopharyngeal carcinoma development. Ras may positively regulate the activity of endogenous RASSF1A [18]. However, in this study we did not find a correlation between RASSF1A and KRAS expression in any lesion evaluated.
KRAS expression was higher in the OSCC group when compared with CG and OLP. Regarding OSCC, we have not found any association between KRAS expression and alcohol abuse, tobacco use, site or gender. However, Tashiro et al., 2020 [19] have reported that KRAS expression was higher in males in OSCC, and its immunohistochemical reactivity was significantly decreased in OSCC compared to that in OLP. In esophageal, the RAS expression was undetectable in precancer lesions, but it was found in SCC [20]. Our results presented a statistical correlation between KRAS overexpression and decreased disease-free survival in OSCC. Increased expression of wild-type KRAS in primary keratinocytes, which have extremely low basal expression levels, resulted in a significant proliferative response and increased cell survival [21]. KRAS is essential for ataxia telangiectasia-mutated interactor (ATMIN) and mediates invasion and migration in HNSCC [22]. A high level of KRAS transcripts was correlated with an advanced TNM stage in patients with HNSCC and was positively associated with ATMIN expression [22].
In summary, RASSF1A expression was similar in the OLD and OSCC groups, suggesting that it plays a critical role in the early stage of OSCC. KRAS expression was higher in OSCC when compared with normal and dysplastic tissues, showing that KRAS expression increases with malignant progression, and its overexpression was associated with worse disease-free survival.
Acknowledgment
We thank Calsavara V for the statistical analysis.
Funding Source
This study was supported by Fundacao de Amparo a Pesquisa do Estado de São Paulo-FAPESP, with undergraduate scholarships to Ribeiro MP (16/18955-5) and Tristão SS (14/20615-2).
References
- van der Waal I. Potentially malignant disorders of the oral and oropharyngeal mucosa; terminology, classification and present concepts of management. Oral Oncol. 2009; 45: 317-323.
- Warnakulasuriya S, Ariyawardana A. Malignant transformation of oral leukoplakia: A systematic review of observational studies. J Oral Pathol Med. 2016; 45: 155-166.
- Nikitakis NG, Pentenero M, Georgaki M, Poh CF, Peterson DE, Edwards P, et al. Molecular markers associated with development and progression of potentially premalignant oral epithelial lesions: Current knowledge and future implications. Oral Surg Oral Med Oral Pathol Oral Radiol. 2018; 125: 650- 669.
- El-Naggar AK, Chan JKC, Rubin Grandis J, Takata T, Slootweg PJ. WHO classification of head and neck tumours. International Agency for Research on Cancer. 2017.
- Choudhari SK, Chaudhary M, Gadbail AR, Sharma A, Tekade S. Oxidative and antioxidative mechanisms in oral cancer and precancer: a review. Oral Oncol. 2014; 50: 10-18.
- Park SH, Kim JJ, Chung JS, Lee SR, Lee GY, Kim HJ, et al. RASSF1A suppresses the activated K-Ras-induced oxidative DNA damage. Biochem Biophys Res Commun. 2011; 408: 149-153.
- Edge S, Byrd DR, Compton CC, Fritz AG, Greene F, Trotti A. American Joint Committee on Cancer. AJCC cancer staging manual. Springer; 7th Edition. 2010.
- Kaminagakura E, Werneck da Cunha I, Soares FA, Nishimoto IN, Kowalski LP. CCND1 amplification and protein overexpression in oral squamous cell carcinoma of young patients. Head Neck. 2011; 33: 1413-1419.
- Herrera Costa F, Narana Ribeiro El Achkar V, Costa V, Paladini I, Kowalski LP, Carvalho YR, et al. Different Expression of Aldehyde Dehydrogenases 1A1 and 2 in Oral Leukoplakia With Epithelial Dysplasia and in Oral Squamous Cell Carcinoma. Appl Immunohistochem Mol Morphol. 2019; 27: 537-542.
- Cao D, Chen Y, Tang Y, Peng X-C, Dong H, Li L-H, et al. Loss of RASSF1A expression in colorectal cancer and its association with k-ras status. Biomed Res Int. 2013; 2013: 976765.
- Elsabah MT, Adel I. Immunohistochemical assay for detection of K-ras protein expression in metastatic colorectal cancer. J Egypt Natl Canc Inst 2013; 25: 51-66.
- Strzelczyk JK, Golabek K, Cuber P, Krakowczyk L, Owczarek AJ, Fronczek M, et al. Comparison of Selected Protein Levels in Tumour and Surgical Margin in a Group of Patients with Oral Cavity Cancer. Biochem Genet. 2017; 55: 322-334.
- Sun J. RASSF1A modulates proliferation-mediated oral squamous cell carcinoma progression. Cancer Cell Int. 2019; 19: 213.
- Guo Y, Stacey DW, Hitomi M. Post-transcriptional regulation of cyclin D1 expression during G2 phase. Oncogene. 2002; 21: 7545-7556.
- Hermann PC, Sancho P, Canamero M, Martinelli P, Madriles F, Gress T, et al. Nicotine promotes initiation and progression of KRAS-induced pancreatic cancer via Gata6-dependent dedifferentiation of acinar cells in mice. Gastroenterology 2014; 147: 1119-1133.e4.
- Wight AJ, Ogden GR. Possible mechanisms by which alcohol may influence the development of oral cancer--a review. Oral Oncol. 1998; 34: 441-447.
- Kaminagakura E, Caris A, Coutinho-Camillo C, Soares FA, Takahama- Junior A, Kowalski LP. Protein expression of CYP1A1, CYP1B1, ALDH1A1, and ALDH2 in young patients with oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2016; 45: 706-712.
- Wang T, Liu H, Chen Y, Liu W, Yu J, Wu G. Methylation associated inactivation of RASSF1A and its synergistic effect with activated K-Ras in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2009; 28: 160.
- Tashiro K, Oikawa M, Miki Y, Takahashi T, Kumamoto H. Immunohistochemical assessment of growth factor signaling molecules: MAPK, Akt, and STAT3 pathways in oral epithelial precursor lesions and squamous cell carcinoma. Odontology. 2020; 108: 91-101.
- Li J, Feng CW, Zhao ZG, Zhou Q, Wang LD. A preliminary study on ras protein expression in human esophageal cancer and precancerous lesions. World J Gastroenterol. 2000; 6: 278-280.
- Hoa M, Davis SL, Ames SJ, Spanjaard RA. Amplification of wild-type K-ras promotes growth of head and neck squamous cell carcinoma. Cancer Res 2002; 62: 7154-7156.
- Li YJ, Lai WT, Chang CC, et al. Ataxia-telangiectasia mutated interactor regulates head and neck cancer metastasis via KRas expression. Oral Oncol. 2017; 66: 100-107.