
Research Article
Austin Diabetes Res. 2016; 1(1): 1001.
Migrational Adaptation and Current Diabetes and Overall Mortality among Various United States and Worldwide Races/Ethnicities
Charles MA1*, Charles RC2, Kane JP3, Hui G4 and Wong ND4
¹Department of Medicine, University of California, San Francisco, USA
²School of Medicine, University of Minnesota, USA
³Department of Medicine and Cardiovascular Research Institute, University of California, USA
4Departments of Epidemiology and Medicine, University of California, Irvine, USA
*Corresponding author: Charles MA, Department of Medicine, University of California, San Francisco, USA
Received: February 17, 2016; Accepted: March 25, 2016; Published: March 29, 2016
Abstract
Out of Africa migrations ~60,000 years ago led to widespread populating of Earth. Ancient migration distances appear related to medical observations including diabetic risk alleles and altitude adaptation. Our objective is to show that ancient migration distance is related to current survival. We measured migration distances and correlated these with diabetes and other US mortality rates and international life expectancy among various racial/ethnic groups. For diabetes there are strong, negative correlations with US mortality rates of ethnic groups and migration distance (r= - 0.70, p< 0.00059), which are confirmed using overall US mortality rates (r=-0.84, p<1x105). There is further confirmation using cardiovascular mortality (r= - 0.86), cancer (r= - 0.71) and sepsis (r= - 0.73). A second data set, World Health Organization 2012 life expectancy data from 192 countries confirm the above US correlations showing a moderate, positive correlation with migration distance (r= 0.41, p < 3 x 10-8 ). R2 analysis indicates that 71% (95% CI=0.48-0.90) of the overall US mortality rate is related to migration distance. When evaluating worldwide life expectancy, migration distance retains 17% (95% CI=0.07-0.27) of its effect on survival. Our data are consistent with migrational adaptation being associated with enhanced US diabetes and other disease survivals for some ethnicities. There is superimposition of substantial influences of current healthy lifestyles/effective health care policies from life expectancy analyses. These interrelationships suggest the dimension of migrational adaptation be further evaluated in the demography of survival research and public health care policy.
Keywords: Out of Africa migration; Diabetes mortality rates; Ancient migration and survival
Introduction
Eighty to ninety percent of diabetic mortality is due to cardiovascular disease. Cardiovascular disease is also overwhelmingly the leading cause of death in the US and other Western societies. Interestingly cardiovascular disease burden or risk factor load appears discordant with mortality rates in certain ethnic groups such as Hispanics and African-Americans. For example Hispanics aged 18 and over or in the Medicare population have increased cardiovascular disease burden, e.g., diabetes, compared to whites [1,2], but paradoxically Hispanics have less diabetes, cardiovascular and overall mortality [2-4]. This phenomenon has been known for over 2 decades and is referred to as the Hispanic paradox [4,5]. Similar paradoxical results have recently been reported in a Veterans Administration population, where African-Americans have increased cardiovascular disease burden (diabetes & hypertension) compared to whites, but similar or reduced cardiovascular mortality [6]. These paradoxical observations led some authors to suggest that genetic factors may be involved.
There are estimates that ~60 thousand years ago (kya) modern humans migrated out of Africa [7]. These migrations are associated with structural genetic variation thought to be modulated by demographic and biological dimensions. For example during migration there is a decrease in genetic diversity consistent with serial founder effects, in which successive migrations, likely caused by local environmental constraints, result in small groups expanding into new areas [8,9]. Further worldwide population genomic structural data are consistent with genetic drift at neutral loci, but accelerated divergence at other loci due to local selection adapting to a broad range of environments creating local phenotypic variation [8,10,11]. Thus new or existing alleles can occur at low frequencies, but in specific environments selective pressure can result in allelic enrichment, which can influence phenotypic expression.
Parallel Genome-Wide Association Studies (GWAS) in regional geographic areas have identified genetic disease risk alleles involved in a myriad of worldwide diseases. These dual sets of above observations of worldwide genetic variance and GWAS should influence our understanding of diseases and thus survivability, and the role that migrational adaptation has in defining these processes. However, diabetes survival has not been evaluated in a worldwide context. Since migration into new and distant geographical areas presents unique environmental stressors engendering adaptation, we now hypothesize that migrational adaptation affects current diabetes survival. The rationale for this hypothesis is that those who migrate farther are likely to experience diverse and accumulating adverse environmental conditions including a) dietary challenges, b) climate and altitude, c) pathogen load and d) hostile humans and animals that evoke genetic alterations influencing disease risk and survival. That diet and climate affect specific allele enrichment related to local adaptation is supported by recent genetic structural studies [11].
Supporting our hypothesis are several apparently disparate, but specific genetically linked phenotypic observations that appear geographically determined from both worldwide and local genetic perspectives. Although there appears to be no reports of specific genetic associations related to mortality in African-American or Hispanic populations, genetic associations for cardiovascular disease burden, such as diabetes have been observed. For example assessment of nDNA alleles related to diseases, e.g., type 2 diabetes, indicate that risk allele frequencies change as migration distance increases out of Africa into Asia and the New World, which appears importantly unrelated to genetic drift [10,12]. As humans migrated into different regions, unrelated groups from different geographical areas independently founded identical alleles more than once, and that risk events appear cumulative and continuous rather than being caused by a single genetic differentiation event [8,10].
Further, evaluation of specific high-risk alleles at the local geographic level permits more genetic and phenotypic detail indicating increased disease risk suggesting that regional, more homogeneous ethnic populations can have uniquely high frequency alleles that appear to drive diseases thought to be polygenic. For example increased diabetes Odds Ratio [OR] of 1.29 in Mexicans is associated with a high frequency (0.50) haplotype, which could account for a 26% diabetes prevalence [13]. This haplotype is rare in Africans and Europeans, present in ~10% of Asians and is attributed to ancient migrations. The extant haplotype’s 5 common Single Nucleotide Polymorphisms (SNPs) originated ~800 kya, and are found in a Neanderthal genome indicating substantial selective pressure to maintain these alleles during migration. Such conservation is possibly related to fuel storage efficiency or innate immunity in response to reduced food availability, since the key gene, SLC16A11, appears primarily active in the liver, and when expressed in HeLa cells results in intracellular triglyceride accumulation [13]. Another risk allele causing premature termination in the TBC1D4 gene, which is related to insulin resistance in skeletal muscle, is found in high frequency (0.17) among Inuits of Greenland, where the diabetes risk OR is a striking 10.3 [14]. This allele is not found in Danish or other Europeans, Chinese or African-Americans. These findings are attributed to natural selection or genetic drift.
Finally, mtDNA haplogroups are differentiated along specific migration routes [15]. mtDNA variants thought to be related to migration and adaptation appear beneficial in certain environments and detrimental in others. For example variant 3394C coding for complex1 protein of the oxidative phosphorylation (OXPHOS) pathway, when present on the M9 haplogroup results in highly coupled OXPHOS due to increased complex1 activity; beneficial for high-altitude Asians [16]. The same 3394C variant on the M9 haplogroup among Asians living at lower altitude is associated with decreased complex1 activity and an increase in Leber hereditary optic neuropathy [16].
Each of the above examples relates to disease risk and appears related to migrational events, whereas herein we present novel correlations with the aim of comparing migration distance with mortality/survival. Our preliminary correlations support the hypothesis that migrational adaptation leads to enhanced current human survival in some races, which suggest that our results may have public health implications.
Methods
Mortality rate acquisition
Several criteria were used to acquire a representative cross section of US survival studies: a) reports were limited to print published national data sets, b) federal governmental, peer-reviewed private and academic reports provide a broad base of mortality rates, c) reports were required to have 4 or more racial/ethnic groups and both genders included, d) multiple age groups were included in overall mortality rate reports, e) mortality rates reported in the last 6 years were used in 7 of 9 studies; all studies were reported within 12 years, f) mortality rate categories used for confirmation of diabetes US mortality rates included only entities that appeared related to potential natural selection, g) mortality rate categories included the top ten causes of US mortality, h) a second data set using a worldwide, national life expectancy table was used to compare with overall US mortality, i) the latter data used average age at death from birth and both genders and j) all mortality rate and life expectancy data are accessible from provided references.
Migration distance acquisition
Migration distances for mortality rate and life expectancy correlations were estimated by determining the length of travel distance along reported, consensus patterns of travel routes (Figure 1) using map distance legends (Table 1). Migration distances are in part underestimates due to sea levels at that time, but the differences between various racial/ethnic groups highlighted herein are of the order of 2-10 fold, thus these estimates appear reasonably valid.
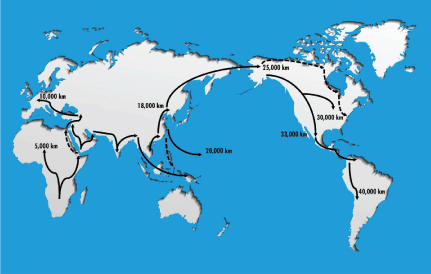
Figure 1: Migration patterns from out of Africa worldwide migrations. Arrows indicate putative pathways used for migration distance estimates, and numbers the
distance traveled in thousands of kilometers. Dashed lines indicate those patterns that remain more controversial. Rationale for migration patterns is detailed in
Methods.
Race/Ethnicity
Distance (Km x 1000)
Black
5
White
10
Asian
18
Asian American/Pacific Islander (AsAm/PI)
18
East Asian/Pacific Islander (A/PI)
18/20 (19)
American Indian/Alaskan Native (AI/AN)
30/25 (27.5)
Native American (NA)
30
Hispanic
33
Mexican-American (Mex-Am)
33
Mexican
33
Central & South America
37/40 (38.5)
Table 1: Estimated migration distances for racial/ethnic groups. Although the routes of travel remain under investigation, there is general agreement with the overall migration patterns and thus approximations of migration distances as shown in Figure 1 and described in detail in Methods.
About 60 kya humans (black race) migrated within (migration distance ~5k Km) and out of Africa to extensively populate Earth. Migrations from Africa along the Nile River valley into Egypt and/ or from southern Africa into Ethiopia and the southern Arabian Peninsula, both led to the Levant [9,17,18] A western migration moved through what is now Turkey ~40kya, and southern, central, western and parts of eastern Europe [19,20]. A northerly migration from the Levant to Georgia, Ukraine and western Russia led to northern and northeastern Europe [21]. These migrations led in part to the Caucasian (white) race; migration distance to western Europe (central France) ~ 10k Km from southern Africa.
The southern African into Ethiopian and southern Arabian Peninsula migration also included a southeastern migration to India along the southern landmass coastline through what is now southern India and Southeast Asia to populate what is now Indonesia, East Malaysia and New Guinea [22,23]. As sea levels were ~300 feet below current levels, the latter three countries islands belonged to a common landmass contiguous with Southeast Asia, known as Sundaland [23]. Northward migration from Southeast Asia via rivers and/or across small land masses to perhaps is what is now Vietnamese coastline led to East Asia ~30 kya [24]. Migration distance: ~18k Km from southern Africa to the North Korean-Chinese border. The East Asian migration populated what is now Taiwan; contiguous with the mainland ~30 kya [24,25]. About 5 kya a Taiwan migration led to the dispersal of East Asian genes into segments of the Philippines, Polynesia, Micronesia and Melanesia [23,25]. Other migrations from Malaysia, New Guinea and the Bismarck islands also led to Pacific Islanders [26], whom today have both Southeast Asian and East Asian ancestry; migration distance to western Polynesia ~ 20k Km. East Asian migrants moved northeasterly across the Bering Sea and Alaska, which was the land mass of Beringia 20-30 kya during the Last Glacial Maximum (LGM). These migrants moved across southern and perhaps the northern coastlines of Beringia into what is now Alaska, leading to the Alaskan Natives [27]; migration distance, ~25k Km. About 15 kya migrations were southeastern from the southern coast of Beringia (Alaska) and/or southern from the north Beringian (Alaskan) coast to western, central and eastern North America leading to the American Indians [27]; migration distance ~30k Km to both Chicago (southern Beringian route) or Washington D.C. (northern Beringian route across what is now Hudson Bay, Canada). Another Pacific coastal, southern migration from Alaska was relatively rapid, requiring only ~ 2k years to reach the southern tip of South America [28]. Migration into the southern part of North America, Central and South America led to the Hispanics; migration distance: ~33k Km to central Mexico, and ~37k and ~40k Km for Central (Panama) and South America (northern Chile) respectively. The Central/South American combined distance is ~38.5Km.
Statistics
Linear regression analyses of mortality rates or life expectancy, modeled as a function of migration distance, were performed providing R2 values and 95% Confidence Intervals (CI) to estimate the proportion of the variation in mortality or life expectancy explained by migration distance. Pearson correlation coefficients were used to show the strength and direction of the relationship of migration distance and mortality rates or life expectancy. For all US mortality rates, data were normalized prior to analyses due to differences in source data, where rates were normalized to rate/1000 persons due to marked rate differences among age groups and genders. For Y-axis correlation analyses, specific racial/ethnic group normalized mortality rates were divided by the white normalized mortality rate, the latter of which was set equal to 1.0, and the resulting fraction for each racial/ethnic group was used (Figure 2 Y-axis, see legend for calculation example and supplements 1-6 for statistical calculations). SAS software was used for the above statistical assessments.
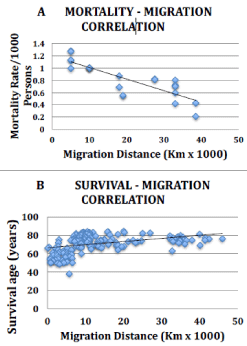
Figure 2: Correlation graphs for migration distance versus various US racial/
ethnic group mortality rates and worldwide life expectancy. A. Overall US
mortality strongly correlates with migration distance. Y-axis coordinates
were determined by calculating the fraction using referenced mortality rates.
For example the published Hispanic/ Mexican-American (Xu, supplement
2, ref 29 for 2007) age-adjusted, overall mortality rate, 546/100,000 was
normalized to 5.46/1000, and then dividing by the normalized white rate of
7.63, the latter of which was set at 1.0 yielding 0.72 for Mexican-American.
See methodology for normalization rationale. B. Life expectancy data,
expressed as mean age at death from birth, were used directly from reference
31. Migration distances were estimated as described in Table 1, Figure 1,
supplement 3A-3C and methods.
Results and Discussion
Several race/ethnicities living in the US, including black, white and East Asian/Pacific Islander, Alaskan Native/American Indian and various Hispanic subgroups have been evaluated for current survival rates, and include a broad spectrum of ancient migration distances (Figure 1, Table 1). Since information from diabetes was the first to suggest migrational adaptation, we formally examined diabetes mortality rates in various ethnicities/races. In a Medicare population [2] adjusted for age and gender, mean diabetes-related mortality rates/1000 for blacks (94.1) exceeded whites (91.9) in the five years evaluated from 1994-2001, but the differences were modest. Substantially lower mean death rates were observed for Hispanics (74.6) and Asians (68.4) in the same study. See Supplement 1 for each year’s data. There is a strong, inverse relationship with overall diabetes mortality (combined for all years reported, supplement 1) and migration distance (r = - 0.70, p<0.00059). These data were consistent with a relationship of migration distance and mortality, thus we sought more definitive data by evaluating overall mortality rates in the general population. To determine if overall mortality rate differences exist between these diverse racial/ethnic groups, we assessed age-adjusted mortality rate/1000 persons. The elderly US population Medicare records [2], (supplement 2) indicate a trend of decreasing mortality starting with blacks (61.0), then whites (54.6), Asians (47.5) and Hispanics (39.2) in 1994; a similar trend was observed in 2001 except that Asians had the least mortality (37.1 vs. 43.3 for Hispanics), and there was no change in white(53.9) or black (61.1) rates. More recent data from 2007 [29] and 2009 [30] from all US individuals (supplement 2) showed mortality rates highest among blacks (9.79; 9.43 for the 2 years), followed by whites (7.63; 7.45), American Indian/Alaskan Native (6.27; 6.03), Hispanics (5.46; 5.19), Asian subgroup (East Asian/Pacific Islanders, 4.15; 4.13) and finally a Hispanic subgroup (Central and South American, 3.26; n/a). Similar studies of 25yrs and older US adults, but using interviews and death certificates between 1997-2004 [4], were restricted to blacks, whites and Hispanics, and again showed decreasing mortality moving from blacks or whites to Hispanics (Supplement 2). In the latter study there was also a markedly decreasing mortality rate/1000persons for Mexican-Americans born in the US (7.30), compared to those born in Mexico (5.15) or Central and South America (2.59). Remarkably correlation analyses between racial/ethnic mortality rates and migration distances reveals a strong, inverse relationship (r = -0.84, p = 1x10-5) (Figure 2A, Supplement 2). R2 is 0.71 (95% CI=0.48-0.90) indicating that migration distance accounts for ~71% of the overall US racial/ethnic mortality rate differences. Using a different statistical model from the latest World Health Organization (WHO), 2012 life expectancy from birth table from 192 countries [31], we found a moderate positive correlation with migration distance (r = 0.41, p < 3x10-8) (Figure 2B, Supplement 3A-3C). R2 is 0.17 (95% CI=0.07-0.27) indicating that migration distance accounts for ~17% of worldwide life expectancy.
Similar results confirm those reported above for diabetes and overall mortality when comparing individual disease-related US mortality rates with the same racial/ethnic groups. For example, in the elderly (65 and older) US population in 1999 [3], cardiovascular disease (heart disease and stroke) by far the leading cause of US mortality/1000 persons, was highest in black males (25.2), followed by white (23.1), Asian/Pacific Islander (14.5), Hispanic (14.3) and Native American (14.1). Similar results were observed for females (supplement 4). For both genders there is a strong, inverse relationship with cardiovascular mortality and migration distance (r = -0.86, p<0.0013) as shown in supplement 4.
To further confirm diabetes and overall US mortality rates using different data sets, cancer survival data were examined, which may in part be related to innate immunity. Cancer mortality rates, the second leading US cause of death, derived from 2002-2006 [32], (supplement 5), indicate that males have the highest rates in blacks (3.04 deaths/1000) followed by whites (2.27), American Indian/ Alaskan Native (1.83), Hispanic (1.55) and Asian American/Pacific Islander (1.35). In the latter study similar results were reported in females [32]. Further cancer mortality data evaluating subjects with non-small cell lung carcinomas (supplement 5) indicate that blacks have the poorest survival (hazard ratio 1.09) followed by whites (1.00 reference category), US born Hispanics (0.85) and non US born Hispanics (0.87) as assessed in 2009 for both genders [33]. There is a strong, inverse relationship with all the above cancer mortality rates and migration distance (r = -0.71, p<0.033).
Pathogen load/innate immunity were assessed using the septicemia surrogate. Septicemia mortality rate/1000 elderly persons in 1999 [3], the ninth leading cause of US deaths, showed the highest death rate in black men (1.50) followed by white (0.66), Native American (0.57), Hispanic (0.47) and Asian/Pacific Islander (0.44). Similar results were observed in females (supplement 6), and for both genders there is a strong, inverse relationship with septicemia mortality rates and migration distance (r = -0.73, p<0.018).
We have presented novel data of current diabetes and overall US mortality rates and worldwide longevity consistent with the hypothesis that migration distance is strongly associated with survival. Potential genetic links to disease-specific death risk led us to the mechanistic concept of migrational adaptation being related to current survival. Public health care policy appears relevant, since Asians and Hispanics have enhanced survival for all measured indicators (>50% reductions for some indicators), when compared to blacks and whites leading us to suggest the dimension of migrational adaptation be further evaluated in the demography of public health care policy for diseaserisk, actual prevalence and survival.
Strengths and limitations
Although correlation analyses are not suitable for addressing causality, our novel migrational adaptation correlations are strengthened by prior studies more directly addressing genetic structure and migration (see Introduction). For example specific disease risk alleles for diabetes (SCL11A16, TBC1D4), longevity alleles in CETP and apo E genes [34-36] and the Hispanic paradox appear related to migration patterns and/or to specific geographic areas (see below). The US may provide unique data for migration distance-related phenomena because a) there are substantial populations of diverse races/ethnicities, b) reasonably reliable mortality statistics exist for diverse races/ethnicities, c) population lifestyles are biased for less exercise and higher caloric intake, which could amplify the signal for population-based prevalence rates, d) medical care is relatively poor for chronic diseases, e.g., diabetes [37], perhaps amplifying population-based mortality rates, thus creating more apparent differences between races/ethnicities and e) social, cultural and ethnic disparities have been well described in multiple races/ethnicities, and thus conceptually can be integrated into our conclusions.
It would appear ideal to obtain current survival data from an array of distant countries of relatively homogeneous racial/ethnic populations; however a) most of these countries have less than optimal mortality rate collection methods, b) some countries reflect current healthy lifestyles, e.g., smoking abstinence, healthy diets/ exercise and effective health care policies, in which more affluent, smaller countries often have the advantage in life expectancy and c) some reflect current poor hygiene, malnutrition and endemic and lethal infectious diseases, where less affluent countries often have a disadvantage in life expectancy. Despite these limitations, we use this type of analysis as a secondary instrument, which provided unexpected findings related to public health, when combined with the US mortality rate data (see below).
There are other possibilities of explaining the possible relationship of migrational adaptation to survival. For example gene admixture within specific racial/ethnic categories in the US could affect survival, but the direction of effect would not explain our results. For example ~20% of the African-American genome has non-African genes, e.g., white (European) and East Asian genes, and since the latter groups have less mortality, that would tend to improve black survival, however despite this potential survival upside, blacks remain with the highest mortality [38]. For Hispanics, black or white (European) gene admixture would tend to decrease rather than improve Hispanic survival as described above, yet Hispanics have improved survival. Also duration of US environmental exposure, e.g., to high calorie/low exercise lifestyles, could be related to current survival. Thus whites and blacks appear most exposed; however Hispanics, also born and raised in the US, have enhanced survival. But deeper examinations of these data are, in part, consistent with the duration of exposure concept, in that US-born Mexican -Americans have higher mortality than more recent US immigrants of Mexican or Central and South American descent [4,5]. Finally, despite the above potential confounding variables, the US Centers for Disease Control and Prevention have recently further documented a 24% reduction in Hispanic mortality rates when compared to whites, which is of similar direction and magnitude to our observation of a ~40% mortality rate reduction [1].
Our data also suggest a threshold survival effect for migrational adaptation of ~10k Km. Although there is a clear directional distinction between black and white mortality rates, with only ~5k Km difference in migration distance; the mortality rate magnitude is somewhat similar. Between blacks/whites and Asians/Hispanics, there are clear mortality rate directional and magnitude differences supporting ~10k Km threshold.
Migrational adaptation may explain the hispanic paradox
Our data showing Hispanic subgroups having among the lowest overall and disease-specific mortality rates, despite having a marked increase in death risk due to increased cardiovascular disease prevalence, low socio-economic status and poor access to medical care has been recognized as the Hispanic paradox [4,5]. Our findings suggest that the unusual Hispanic survival is not paradoxical, but in fact related to migrational adaptation. We would expand this new explanation to include East Asian/Pacific Islander and Alaskan Native/American Indian ethnicities.
Longevity
Longevity genes have been described and appear regionally relevant. For example apolipoprotein E and Cholesteryl Ester Transfer Protein (CETP) alleles are associated with human longevity, and the minor allele frequencies occur in 10-15% of individuals of Dutch, Danish and German ancestry, but not in individuals from other European areas [34,35]. Since these proteins appear related to innate immunity, we reasoned that migrational adaptation could be related to longevity via innate immunity [36,39]. Innate immunity also appears involved in cardiovascular disease, cancer and infectious disease. We have recently elucidated the mechanisms of lipid transfer in CETP [40], and similar mechanisms are thought to exist in other CETP family members, which are implicated with innate immunity, the latter of which is important throughout all phases of the human life cycle. We are unaware of worldwide genetic/migration longevity studies.
Genetic implications
The evolution of complex phenotypes is likely maintained by SNPs at multiple gene loci responding to local environmental pressures [41]. In this context, it has been suggested that the two newest selectively evolved human genes are 3000-7000 years old [42]. These observations imply that significant time is required for adaptively selected genes to manifest, whereas purifying selection due to lack of adaptive value, occurs after several hundred years. These concepts may explain why Hispanics and Asians have high prevalence rates of cardiovascular risk burden driven by relatively recent (~150 years) lifestyle patterns coupled with their long standing genetic background of fuel conservation efficiency, possibly related to prolonged and repeated episodes of reduced fuel supplies during migration. These current lifestyles are not yet susceptible to purifying selection due to their recent occurrence and the lethal effects occurring after the procreation stage of life; but nevertheless having lower mortality rates, possibly driven by other unknown alleles also related to migration, e.g., innate immunity.
Although type 2 diabetes risk alleles decrease with migration distance out of Africa [10,12], the current phenotype of increased US cardiovascular disease and diabetes prevalence, in both blacks and Hispanics, is thought to be driven by the local US environment, e.g., decreased exercise and/or increased caloric intake. This environmentally driven high prevalence is also likely gene related, either through diabetes risk alleles (examples SLC16A11, TBC1D4), obesity alleles (possibly IRX3) and the thrifty genotype [13,14,43-45].
The apparent discordance of reduced diabetes risk alleles and elevated diabetes prevalence can be explained by the actions of alleles not included in the worldwide assessment of diabetes risk alleles [10,12]. The genetic backgrounds of current US immigrants and incumbents were determined well over 3- 7 kya, when the newest human alleles were expressed, thus rendering current US race/ethnicspecific social, cultural, and medical access influences (~150 years) of less significance than their genetic background. The Hispanic paradox supports this notion. Further if sudden, new environmental pressures ensue, e.g., alterations in caloric intake/exercise levels, and elevated prevalence occurs in proportion to the background genotypes of the various racial/ethnic populations (especially Hispanics), then one would expect survival rates to be proportional to the newly increased prevalence. Interestingly our observations lead us to speculate that US survival-related genetic background may be dominant to the currently increased disease prevalence, and perhaps some genotypes associated with migrational adaptation are capable of driving survival under extremely adverse genetic and environmental conditions. Thus our hypothesis construction included dietary challenges, but that appears more relevant for prevalence than survival.
Lifestyle/Effective health care policy implications
Our R2 analysis indicates that 71% of the variation in the overall US mortality rate is related to migration distance, consistent with migrational adaptation. This finding suggests other less significant factors also affecting US mortality, e.g., lifestyles, socio-demographic factors and health care policies. Parsimoniously, our worldwide correlation r-values support our hypothesis, but the R2 analysis suggests alternative concepts. Thus migration distance accounts for only 17% of the variation in worldwide life expectancy. These data suggest a substantial influence of factors, e.g., lifestyle, sociodemographic factors or health care policies. It is important to note that the two 95% CIs do not overlap indicating that the proportion of variation explained by migration distance in predicting overall mortality is significantly greater than that predicting life expectancy. Virtually all of the top 10 countries in the 2012 WHO life expectancy data are relatively small and socio-demographically more homogeneous countries when compared to the US, and they have healthy lifestyles and effective, inclusive health care policies suggesting that these dimensions are currently more important than migrational adaptation for these countries. Life expectancy data are consistent with the concept that lifestyle and effective health care policy can, to some extent override migrational adaptation, but that the latter can be dominant to the former in large, diverse countries with relatively poor healthcare outcomes, like the US [37]. In some countries, e.g., Japan and Singapore, migrational adaptation and healthy lifestyles/effective health care policies are combined perhaps leading to their number 1 and 3 rankings in the world [31].
Conclusion
Our data are consistent with migrational adaptation being associated with natural selection of several traits, which may enhance current diabetes and overall US survival for some ethnicities. There is however, superimposition of substantial influences of current healthy lifestyles/effective health care policies from worldwide life expectancy analyses. These important interrelationships suggest the dimension of migrational adaptation be further evaluated in the demography of longevity research and public health care policy. Our data also suggest that within a diverse US population, specific racial/ethnic groups may require reevaluation of risk/survival preventive health care policy.
References
- Leading causes of death, prevalence of diseases and risk factors, and use of health services among Hispanics in the United States. Morbidity and mortality weekly report. Centers for Disease Control and Prevention. 2015; 64: 469- 478.
- McBean AM, Li S, Gilbertson DT, Collins AJ. Differences in diabetes prevalence, incidence, and mortality among the elderly of 4 racial/ethnic groups: whites, blacks, Hispanics, and Asians. Diabetes Care. 2004; 27: 2317-2324.
- Anderson NB, Bulatao RA, Cohen B. Critical perspectives on racial and ethnic differences in health and late life. National Research Council (US) Panel on Race, Ethnicity and Health in Later Life. Washington DC: National Academic Press. 2004.
- Borrell LN, Lancet EA. Race/ethnicity and all-cause mortality in US adults: revisiting the Hispanic paradox. Am J Public Health. 2012; 102: 836-843.
- Fenelon A. Revisiting the Hispanic mortality advantage in the United States: the role of smoking. Soc Sci Med. 2013; 82: 1-9.
- Kovesdy CP, Norris KC, Boulware LE, Lu JL, Ma JZ, Streja E, et.al. Association of race with mortality and cardiovascular events in a large cohort of US veterans. Circulation. 2015; 132: 1538-1548.
- Mellars P, Gori KC, Carr M, Soares PA, Richards MB. Genetic and archaeological perspectives on the initial modern human colonization of southern Asia. Proc Natl Acad Sci USA. 2013; 110: 10699-10704.
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008; 319: 1100-1104.
- Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc Natl Acad Sci U S A. 2012; 109: 17758-17764.
- Corona E, Chen R, Sikora M, Morgan AA, Patel CJ, Ramesh A. et. al. Analysis of the genetic basis of disease in the context of worldwide human relationships and migration. PLoS Genet. 2013; 9: e1003447.
- Hancock AM, Witonsky DB, Ehler E, Alkorta-Aranburu G, Beall C, Gebremedhin A. et. al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Nat Acad Sci. 2010; 107:8924-8930.
- Chen R, Corona E, Sikora M, Dudley JT, Morgan AA, Moreno-Estrada A. et. al. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. PLoS Genet. 2012; 8:e1002621.
- SIGMA Type 2 Diabetes Consortium, Williams AL, Jacobs SB, Moreno- Macías H, Huerta-Chagoya A, Churchhouse C, Márquez-Luna C, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014; 506: 97-101.
- Moltke I, Grarup N, Jorgensen M, Bierregaard P, Treebak JT, Fumagalli M, et. al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014; 512:190-193.
- Wallace DC. Bioenergetics in human evolution and disease: implications for the origins of biological complexity and the missing genetic variation of common diseases. Philos Trans R Soc Lond B Biol Sci. 2013; 368: 20120267.
- Ji F1, Sharpley MS, Derbeneva O, Alves LS, Qian P, Wang Y, et al. Mitochondrial DNA variant associated with Leber hereditary optic neuropathy and high-altitude Tibetans. Proc Natl Acad Sci U S A. 2012; 109: 7391-7396.
- Lawler A. Human evolution. Did modern humans travel out of Africa via Arabia? Science. 2011; 331: 387.
- Osborne AH, Vance D, Rohling DL, Barton N, Rogerson M, Fello N. A humid corridor across the Sahara for the migration of early modern humans out of Africa 120,000 years ago. Proc Nat’l Acad Sci. 2008; 105: 16444-16447.
- Deguilloux MF, Leahy R, Pemonge MH, Rottier S. European neolithization and ancient DNA: an assessment. Evol Anthropol. 2012; 21: 24-37.
- Fu Q, Rudan P, Pääbo S, Krause J. Complete mitochondrial genomes reveal neolithic expansion into Europe. PLoS One. 2012; 7: e32473.
- Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, et al. The archaeogenetics of Europe. Curr Biol. 2010; 20: R174-183.
- Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005; 308: 1034-1036.
- Jinam TA, Hong LC, Phipps ME, Stoneking M, Ameen M, Edo J, et al. Evolutionary history of continental Southeast Asians: “early train” hypothesis based on genetic analysis DNA of mitochondrial and autosomal data. Mol Biol Evol. 2012; 29: 3513-3527.
- HUGO Pan-Asian SNP Consortium, Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC. Mapping human genetic diversity in Asia. Science. 2009; 326: 1541-1545.
- Mirabal S, Cadenas AM, Garcia-Bertrand R, Herrera RJ. Ascertaining the role of Taiwan as a source for the Austronesian expansion. Am J Phys Anthropol. 2013; 150: 551-564.
- Soares P, Rito T, Trejaut J, Mormina M, Hill C, Tinkler-Hundal E, et al. Ancient voyaging and Polynesian origins. Am J Hum Genet. 2011; 88: 239-247.
- O’Rourke DH, Raff JA. The human genetic history of the Americas: the final frontier. Curr Biol. 2010; 20: R202-207.
- Bodnar M, Perego UA, Huber G, Fendt L, Rock AW, Zimmermann B, et al. Rapid coastal spread the first Americans: Novel insights from South America’s Southern Cone mitochondrial genomes. Genome Res. 2013; 22: 811-820.
- Xu J, Kochanek KD, Murphy SL, Tejada-Vera B. Deaths: final data for 2007. Natl Vital Stat Rep. 2010; 58: 1-19.
- Minino AM. Death in United States, 2009. National Center for Health Statistics Data Brief. 2011; No. 64. HHS, Centers for Disease Control and Prevention.
- Life expectancy data by country. World Health Organization. 2012.
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010; 60: 277-300.
- Saeed AM, Toonkel R, Glassberg MK, Nguyen D, Hu JJ, Zimmers TA, et al. The influence of Hispanic ethnicity on nonsmall cell lung cancer histology and patient survival: an analysis of the Survival, Epidemiology, and End Results database. Cancer. 2012; 118: 4495-4501.
- Beekman M, Blanché H, Perola M, Hervonen A, Bezrukov V, Sikora E, et al. Genome-wide linkage analysis for human longevity: Genetics of Healthy Aging Study. Aging Cell. 2013; 12: 184-193.
- Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer’s disease to AIDS. J Lipid Res. 2009; 50: S183-188.
- Stark Casagrande S, Fradkin JE, Saydah SH, Rust KF, Cowie CC. The prevalence of meeting A1C, blood pressure, and LDL goals among people with diabetes, 1988-2010. Diabetes Care. 2013; 36: 2271-2279.
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et. al. The genetic structure and history of Africans and African Americans. Science. 2009; 324: 1035-1044.
- Cazita PM, Barbeiro DF, Moretti AIS, Quintao ECR, Soriano FG. Human cholesteryl ester transfer protein expression enhances the mouse survival rate in an experimental systemic inflammation model: a novel role for CETP. Shock. 2008; 30: 590-595.
- Zhang LF, Yan F, Zhang S, Lei D, Charles MA, Cavigiolio G, et al. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat Chem Biol. 2012; 8: 342-349.
- Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, et al. The role of geography in human adaptation. PLoS Genet. 2009; 5: e1000500.
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010; 329: 75-78.
- Grarup N, Sandholt CH, Hansen T, Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014; 57: 1528-1541.
- Tung YC, Yeo GS, O’Rahilly S, Coll AP. Obesity and FTO: Changing Focus at a Complex Locus. Cell Metab. 2014; 20: 710-718.
- Neel JV. The “thrifty genotype” in 1998. Nutr Rev. 1999; 57: S2-9.
- Soerensen M, Dato S, Tan Q, Thinggaard M, Kleindorp R, Beekman M, et al. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr). 2013; 35: 487-500.