
Research Article
J Dis Markers. 2015;2(2): 1024.
Tetraspanin Expression Profile in Soft Tissue Sarcomas
Katia C Carvalho¹*, Isabela W Cunha², Rafael M Rocha³, Marcilei E Buim³, Mariana Maschietto³,Yukie S Kuwabara³, Luiz F L Reis4 and Fernando A Soares2,3
¹Laboratório de Ginecologia Estrutural e Molecular (LIM-58), Disciplina de Ginecologia, Hospital da Clinicas daFaculdade de Medicina da Universidade de Sao Paulo,Brazil
²Departamento de Anatomia Patológica, AC Camargo Cancer Center, Sao Paulo, Brazil
³Centro Internacional de Pesquisas (CIPE), AC Camargo Cancer Center, São Paulo, Brazil
4Hospital Sirio-Libanes, São Paulo, SP, Brasil
*Corresponding author: Katia Candido Carvalho,Obstetrics and Gynecology Department - FMUSP, Sao Paulo University, Cerqueira Cesar - Sao Paulo/SP, Brazil.Cep. 01246-903
Received: April 14, 2015; Accepted: May 27, 2015; Published: May 29, 2015
Abstract
Sarcomas are rare malignant tumors rich in stromal and extracellular material; however, the features involved in their adhesion and cell migration are unclear. Moreover, little is known about molecular alterations, which could contribute to their malignant transformation and biology. We have previously observed a lack of correlation in tetraspanin expression module on sarcomas and a poorer prognosis, using a cDNA microarray platform. Tetraspanins are proteins, which are involved in several cellular processes and their profile, or role in sarcomas is unknown. Our aim was to validate cDNA microarray data and to evaluate the correlation of tetraspanins expression profile with clinical pathological parameters. Protein and transcript expression were evaluated by immunohistochemistry (227 samples) and quantitative real time - PCR (47 samples). Results showed a significant downregulation of CD81 and CD37 transcripts in high grade and metastatic tumors. CD63 showed the highest frequency of cytoplasm protein expression, showing a trend of association with poor survival (p=0,053). CD9 protein was observed, mainly in the membrane of leiomyosarcomas and pleomorphic sarcomas. Taken together, our results indicate that CD9, CD81 and CD82 does appear to play an important role in soft tissue sarcomas prognosis and their higher expression correlates with less aggressive forms of behavior. The role of CD63 protein needs further investigation, due to its high frequency and intensity of expression in STS.
Keywords: Sarcomas; Tetraspanins; qRT-PCR; Immunohistochemistry; Prognosis
Introduction
Sarcomas are malignant tumors of the connective tissue and can be classified in different histological types [1]. In their early stages, patients are asymptomatic, because soft tissues are relatively elastic. This enables tumor growth without presenting alterations or sequelae [2]. Although these neoplasms are relatively rare, patients present high rates of mortality and morbidity [3]. Generally, sarcomas are histologically classified according to their differentiated tissue types (eg. leiomiosarcoma, liposarcoma, osteosarcomas and others) [1,2]. Diagnosis is based on the histological type and tumor grade [2,4]. Whilst tumor size and histological features are the best prognostic factors available for mesenchymal tumors, very little is known about their molecular alterations, which could contribute to our understanding of the cells origins, malignant transformations, and tumor biology of sarcomas [4]. Several previous works have shown an aberrant tetraspanin (TSPAN) expression in cancer, but do not associate these protein families with sarcomas’ clinical or pathological features.
These proteins are integral membrane proteins, which are characterized by the presence of four transmembrane domains. They belong to a large family, in which there are at least 33 members, expressed all cell types except for red blood cells [5]. Countless studies have been done which demonstrates the involvement of TSPAN in several physiological processes. Including tissue differentiation, cell proliferation, cell-matrix adhesion, cell migration, viral-induced syncytium formation, fusion processes, signal transduction and cellular activation [6]. Experimental and clinical studies have shown the importance of TSPAN in tumor biology and behavior [7,8].
Studies have analyzed human tumors from different primary sites and shown the importance of TSPAN in cancer progression, metastasis and prognosis. The downregulation of these proteins, is in general, associated with the development of metastasis or the loss of cell adhesion and neoplastic cell invasion [9,10]. However, a pro-tumoral role has also been identified with some TSPAN. The transfection of CD151 into cancer cells demonstrated an enhancement of the cell motility, invasion and metastatic potential in vitro of human colon cancer cells, glioblastoma and fibrosarcoma [11]. Research also demonstrated that a spliced variant of CD82, obtained by the excision of exon 7, was associated with an increase in cell motility, loss of cell adhesion, tumor growth and metastasis occurrence [12].
Although the importance of TSPAN in a wide range of epithelial tumors is well known, their role in soft tissue sarcomas (STS) has not been investigated. In a preliminary study, with 73 histologically different soft tissue tumors in a cDNA microarray platform, (containing 4.608 ORESTES sequences); we found a lack (change) of linear correlations between TSPAN gene pairs when considering tumor characteristics and gene expression profiles. CD9, CD37, CD63, CD81 and CD82 presented significant differences in their fold of expression. We aimed our investigation therefore, at the gene and protein expression of CD9, CD37, CD63, CD81 and CD82 and to evaluate the correlation of their expression profile with clinical pathological parameters.
Material and Methods
Tissue samples
Tissue samples were obtained from a total of 227 patients undergoing operative procedures on primary tumor sites, between 1973-2006. All tissue samples were obtained from the Tumor Bank and Paraffin Block archive, in the Department of Anatomic Pathology of the A.C. Camargo Cancer Center, São Paulo, Brazil. Hematoxylin and eosin stained slides were reviewed by two pathologists (IWC and FAS) and classified according to the criteria of 2013 World Health Organization (WHO). Among the available systems for sarcomas graduation, we used the French Fédération Nationale des Centres de Lutte Contre le Câncer (FNCLCC) and the National Cancer Institute (NCI). The Institutional Review Boards of the A.C. Camargo Cancer Center (Study Number 1031/08) approved this study and preoperative informed consents were obtained from all patients.
The follow-up of records of patients, for a period of more than sixty months, and all clinical and pathological data was obtained from patient archives. Sex, ethnicity, age, treatment, tumor size and staging, metastasis, recurrence, survival and other clinical features, were evaluated. For purposes of statistical analysis, we grouped intermediate and high histological grade tumors, and moderate and high protein expressions. When data analysis was performed separately, no significant differences were discovered, probably due the low number of samples for some histological types of tumors.
cDNA microarray, mathematical analysis and quantitative real time (qRT)-PCR
The cDNA microarray analysis was performed as described previously [13]. To analyze the relationship of TSPANs between 73 different frozen samples, we mapped a microarray platform containing 4.608 ORESTES sequences (Ludwig Institute chip [13,14] in order to find the presence of these genes on the chip. Detailed descriptions are available at Gene Expression Omnibus data repository under accession number GPL1930, and the accession number for raw data is GSE14541 (https://www.ncbi.nlm.nih.gov/ projects/geo). We performed a relevance networks analysis, which enables the visualization of the correlations between molecules under different conditions. A Fisher’s Z transformation was used to obtain the difference in significance between the correlations undergoing different analyses [15]. The initial chosen criteria for evaluation were: 1) metastatic versus non-metastatic tumors (after a five-year period of patient follow-up), 2) lower versus higher histological grade tumors (at diagnosis stage), and 3) patient status (death due to cancer or alive, free from cancer disease, after a five-year period of follow-up).
47 frozen samples of STS (previously evaluated by cDNA microarray) were investigated for the CD9, CD37, CD63, CD81 and CD82 gene expression; by qRT- PCR (Table 1). RNA was obtained using the Trizol method (Life Technologies, Carlsbad, CA) and firststrand cDNA was synthesized with High-Capacity cDNA Reverse Transcription Kit (Life Technologies). The synthesis was performed from 2 μg of total RNA samples, following manufacturer suggestions. The qRT-PCR reactions were performed using applied biosystems 7900HT Fast Real-Time PCR System (Life Technologies).
Histological tumor type
Immunohistochemical Number of samples
cDNA Microarray Number of samples
Real – Time PCR Number of samples
Pleomorphic sarcoma
45
17
14
Synovial sarcoma
50
19
14
Liposarcoma
35
05
03
Fibrosarcoma
11
06
03
Leiomyosarcoma
52
19
09
Alveolar sarcoma
02
02
02
MPNST
04
05
02
Mixofibrosarcoma
12
-
-
Others types
16
-
-
Total of samples
227
73
47
Table 1: Histological types of the samples evaluated in this study.
The PCR assays were performed as previously described [16]. The assays (Life Technologies) selected for our target genes were: CD9 (Hs 00233521_m1), CD37 (Hs 01099648_m1), CD63 (Hs 00156390_ m1), CD81 (Hs 01002167_m1) and CD82 (Hs 00356310_m1). β-actina (4326315E), GAPDH (4326317E), B2M (4326319E), HPRT (4326321E), TBX (4326322E) were used for housekeeping controls.
Quantitative real-time PCR reactions were performed in triplicate and we used a pool of RNAs samples obtained from 15 different human cell lines as reference [13]. Analysis was performed using the ΔΔCT method, using the SDS 3.0 software (Life Technologies).
Tissue microarray (TMA) and Immunohistochemistry
For the immunohistochemical (IHC) evaluation of protein expression, two paraffin blocks of Tissue microarray (TMA) containing 227 paraffin-embedded STS samples (including the samples evaluated by cDNA microarrays and qRT-PCR) were constructed as previously described [17]. All antibody dilution was standardized and evaluated in conventional slides for evaluation of TMA viability and protein analysis in our tissues. The slides contained representative samples of the STS tissues. We decide to perform IHC in TMA slides because the evaluation of all 227 samples could be carried out simultaneously. The same conditions were applied in all cases, decreasing the variation or bias on the test results. The immunohistochemistry test was also performed on TMA slides, which contained the normal soft tissue, collected from autopsies.
The IHC tests for CD9, CD 37, CD63, CD81 and CD82 were carried out as described previously [18]. The immunostaining was evaluated independently by two pathologists (IWC and FAS), and scored semi-quantitatively by assessing the intensity of staining and frequency of positive tumor cells [17,18]. The intensity of staining was divided in four groups in terms of cytoplasmic and/or membrane staining: 0 = negative; 1 = weak; 2 = moderate; and 3 = strong. The frequency scores of immunostained cells were ranked into four groups: 1) less than 10%; 2) 10% to 50%; 3) 50% to 90%; 4) more than 90% of tumor cells. A combined score ranging from 0 to 12 was calculated by the multiplication of (intensity scores) and (frequency scores), defining four groups: negative (scores 0-2), weakly positive (scores 3-4), moderate (scores 5-8) and strongly positive (scores 9-12). Two groups were studied for Statistical analysis: Negative (0- 4, including negative and weakly positive) and positive (moderate and strongly positive). In the case of any discrepancy arising between scores determined by the two observers (FAS and IWC), the average score was considered.
Statistical analysis
Fisher’s exact test was used to compare the clinical pathological data of patients (age, sex, ethnic, histological tumor type and size, histological grade, metastasis, local recurrence and survival) together with the IHC and qRT-PCR expression data. The χ2 test was used to evaluate the differences between the groups of categorical variables. Overall and Recurrence Free Survival rates were calculated using the Kaplan–Meier method, based on a follow-up period of 5 years for all patients included in this study. A Log rank test was used in the multivariate analysis. All calculations were done using the Graph Pad Prism 5.0 statistical software (La Jolla, CA, USA) and SPSS for Windows (SPSS Inc. Chicago, IL, USA), with a confidence interval of p≤0.05.
Results
Mathematical analysis of cDNAs microarray and tetraspanins gene expression
Based on data which demonstrates patient age range as being between 13 to 86 years (mean 40 ± 7, 0 years) together with a followup period of more than five years (1983 – 2006). We observed the following results:
There is no gender predominance (49% female and 51% male) in our samples. 73% of patients were Caucasian, 22% were black and 5% unknown. Tumor sizes ranged from 1.3 cm to 13 cm, metastasis and local recurrence were observed in 53% and 35% of cases, respectively. From the patients studied 36% died from cancer, 45% are still alive (30% are cancer free and 15% are still living with cancer) and 19% remain unknown (patients were not found). The supplementary material shows data by histological tumor type and statistical analyses.
The data from the cDNA microarrays were analyzed mathematically with regards to the TSPAN web presence and expression. Results showed a lack, or change, in linear correlation of several of the TSPAN gene pairs. Data from cDNA microarray analyses were selected for validation by qRT-PCR and immunohistochemistry. Our chip was used in the human cancer genome project and represents approximately 4,800 genes involved in carcinogenesis. We searched for the gene names using the NCBI (National Center for Biotechnology Information) gene bank for reference.
A transcriptional profile of the molecules evaluated in all tumor types can be seen in Figure 1. CD81 showed a differential expression between high and low histological grade tumors. High-grade tumors showed a downregulation of CD81 transcriptional level (p=0.0121, Figure 2A). Similar results were identified between metastatic and non-metastatic tumors. Regarding CD37, the expression level was significantly higher in non-metastatic when compared to the metastatic tumors (p=0.0315, Figure 2B).
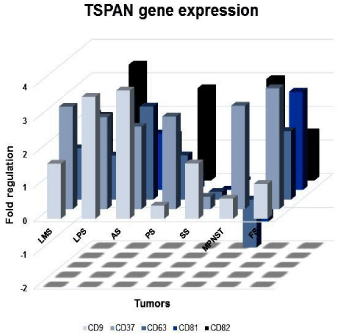
Figure 1: Profile of CD9, CD37, CD63, CD81 and CD82 tetraspanins gene
expression. Graphic representation of CD9, CD37, CD63, CD81 and CD82
tetraspanins transcriptional expression among different histological types of
tumors evaluated by qRT-PCR. LMS: Leiomiosarcomas; LPS: Liposarcomas;
AS: Soft part alveolar sarcomas; PS: Pleomorphic Sarcomas; SS: Synovial
Sarcomas; MPNST: Malignant Peripheral Nerve Sheath Tumors; FS:
Fibrosarcomas.
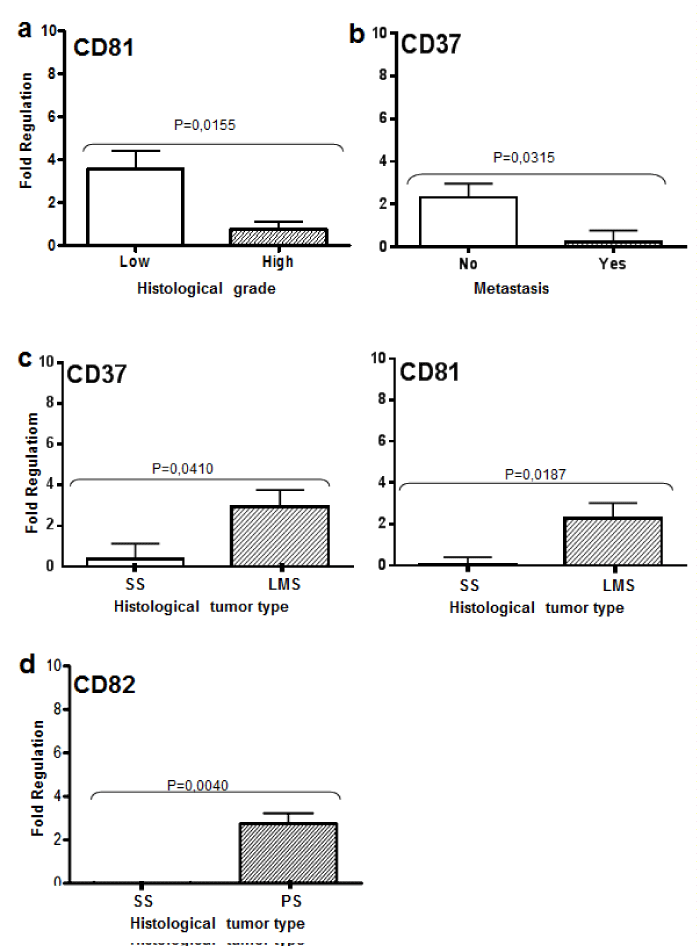
Figure 2: Tetraspanins expression according to clinicopatological features.
In the graph A, CD81 relative expression values obtained considering high
(High) and low (Low) grade tumors by qRT-PCR (p<0, 05). B, CD37 relative
expression values obtained considering presence (Yes) or absence (No) of
metastasis. C, CD82 relative expression values obtained for synovial sarcoma
(SS) and pleomorphic sarcoma (SP). D, Values of relative expression of
CD81 and CD37 obtained in synovial sarcoma (SS) and leiomyosarcoma
(LMS) by qRT-PCR. Statistical significance is indicated (p value).
When comparing histological types, CD81 and CD37 were upregulated in leiomyosarcomas compared to synovial sarcomas (p=0.0186 and p=0.0409, respectively) (Figure 2C). CD82 was upregulated in pleomorphic sarcomas when compared with synovial sarcomas (p=0.004) (Figure 2D). All TSPAN transcripts showed downregulation in synovial sarcomas.
No significant differences (p>0.05) were obtained for transcriptional expression with CD9, CD37, CD63, CD81 and CD82 tumors with regards to age, gender, ethnicity, local recurrence and survival.
CD9, CD37, CD63, CD81 and CD82 protein expression
CD63, CD81, CD82 showed both membrane and cytoplasmic reactivity. CD9 showed exclusively membrane staining. Immunohistochemical (IHC) analysis showed a strong expression of CD63 protein in 71% (32 out of 45 samples) of pleomorphic sarcomas, 66% (33 out of 50 samples) of synovial sarcomas and 65% (34 out of 52 samples) of leiomyosarcomas (Figure 3A-C). Alveolar soft part sarcomas (2 out of 2 samples), malignant peripheral nerve sheath tumors (MPNST) (2 out of 4 samples) and fibrosarcomas (3 out of 11 samples) showed positive CD63 staining. Liposarcoma did not show CD63 expression. CD37 protein expression was negative in all evaluated tumors. Table 2 and Figure 4 show the percentage and scores of IHC positive samples.
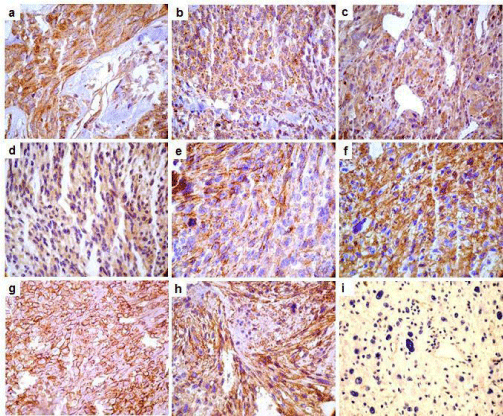
Figure 3: Photomicrography representation of CD9, CD63, CD81 and CD82 tetraspanins in samples of Soft Tissue Sarcomas. Immunohistochemical reaction
showing samples stained for tetraspanins (200 and 400X). CD63 positive reactions are showed in the panels: A (sample of pleomorphic sarcoma), B (synovial
sarcoma) and C (leiomyosarcoma). Panel D shows a positive sample for CD81 from synovial sarcoma. Panel E shows a CD82 positive sample in a MPNST. CD9
positives samples are presented in the panels: F (leiomyosarcoma), G (pleomorphic sarcoma) and H (synovial sarcoma). Panel I shows negative sample for CD63.
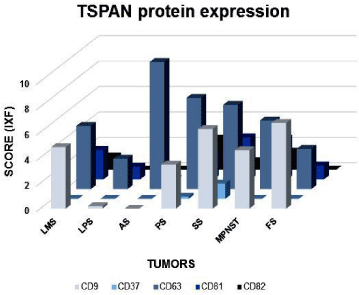
Figure 4: Profile of CD9, CD37, CD63, CD81 and CD82 tetraspanins protein
expression. Graphic representation of CD9, CD37, CD63, CD81 and CD82
tetraspanins IHC positive expression (strong and moderate) among different
histological types of tumors. LMS: Leiomiosarcomas; LPS: Liposarcomas;
AS: Soft part alveolar sarcomas; PS: Pleomorphic Sarcomas; SS: Synovial
Sarcomas; MPNST: Malignant Peripheral Nerve Sheath Tumors; FS:
Fibrosarcomas.
Histological tumor type
Cases / Total samples (percentage of positive samples)
CD9
CD63
CD82
CD81
Pleomorphic sarcoma
10/45 (22%)
29/45 (71%)
5/45 (11%)
6/45 (13%)
Synovial sarcoma
11/50 (22%)
33/50 (66%)
1/50 (2%)
5/50 (10%)
Liposarcoma
10/35 (28%)
-
-
-
Fibrosarcoma
3/11 (28%)
3/11 (28%)
-
-
Leiomyosarcoma
15/52 (29%)
34/52 (65%)
-
8/52 (15%)
Alveolar sarcoma
-
2/2 (100%)
-
-
MPNST
3/4 (75%)
2/4 (50%)
1/4 (25%)
1/4 (25%)
Mixofibrosarcoma
2/12 (17%)
4/12 (33%)
-
-
Others types
1/16 (6%)
8/16 (50)
-
-
Total of positive cases
55
115
7
20
Table 2: Number of positive cases (moderate and strong staining) for the CD9, CD63, CD81 and CD82 tetraspanins according to different histological types of soft tissue sarcomas evaluated by IHQ.
IHC analysis for CD81 and CD82 showed weaker intensity of staining compared to other TSPAN (Figures 3D and 3E, respectively). Only 11% (5 of 45 samples) of evaluated pleomorphic sarcoma samples showed CD82 expression, followed by 2% (1 of 50 samples) of synovial sarcoma and 25% (1 of 4 samples) of MPNST. CD81 was found in 15% (8 out of 52 samples) of Leiomyosarcoma, 10% (5 of 50 samples) of synovial sarcoma and 13% (6 of 45 samples) of pleomorphic sarcoma. No expressions of CD81 or CD82 were detected in liposarcoma, alveolar soft part sarcomas, fibrosarcoma and myxofibrosarcoma. In addition, leiomyosarcoma did not show any positive staining for CD82.
CD9 showed a moderate intensity pattern of expression in 29% (15 out of 52 samples) of leiomyosarcomas (Figure 3F), 28% (10 out of 35 samples) of liposarcomas, 22% (10 out of 45 samples) of pleomorphic sarcomas (Figure 3G) and 22% (11 out of 50 samples) of synovial sarcomas (Figure 3H). Alveolar soft part sarcomas showed negative for this protein expression (Figure 3I).
No significant association between clinical pathological data and IHC results were found (p>0.05), although we found a tendency of expression similar to that described for mRNA. CD63 showed a marginal p value (p=0.053; Figure 5) associated with disease-free survival.
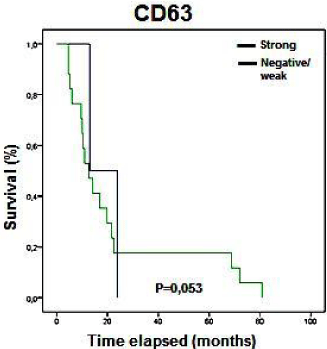
Figure 5: Kaplan-Meier curves showing the disease free survival rates
(months) of patients with STS in relation to CD63 expression status analyzed
by IHC. Only marginal p values were observed (0.053).
Concerning to correlation between protein and gene expression, Spearman tested showed positive correlation for CD9 (in the SS, PS and LMS samples) and CD63 (in the SS samples).
Discussion
This study provides the first available evidence of a TSPAN expression profile in STS, both at protein and mRNA levels. Our aims were to verify these molecule expressions in STS, as some histological types of tumor had shown an increased aggressiveness compared to others. In addition, it was also known that a reduced or lack of TSPAN expression was commonly found in metastatic lesions and poorer prognosis carcinomas [19-22]. In this study, immunohistochemistry was performed in 227 FFPE (Formalin Fixed Paraffin Embedded) tissue samples and their transcript expression was assessed in 47 samples of frozen tissue. Considering that STS are rare tumors, these numbers are significant and our results contribute greatly to the understanding of these neoplasms.
We were able to detect and evaluate most of the selected TSPAN. Although immunohistochemical results did not show a statistically significant association with evaluated clinical parameters, the protein levels demonstrated a higher trend of expression in tumors with a good prognosis (without metastasis or local recurrence, lower histological grade and disease-free patients).
The histological grade of differentiation is very important in STS diagnosis, since sarcoma staging is based on tumor grade, size and location [1,2]. We observed that lower grade tumors presented higher quantities of CD81 transcript. This TSPAN, also called TAPA-I, has been described as a target for the anti- proliferative monoclonal antibody [23]. CD81 is an endothelial lateral junction component, which is implicated in the regulation of cell motility and adhesion in lymphocytes. It may also induce astrocytes cell arrest after brain damage [24]. Over expression of CD81 also reduces viability and motility in multiple myeloma cell lines [5,7,22]. Our results corroborate with the data literature, since several types of poor prognosis carcinomas showed a lack of CD81 expression [16,19,22]. This suggests the same prognosis value in sarcomas.
In this study, metastatic tumors showed a lower transcript expression of CD37 when compared to non-metastatic tumors. The expression of CD37 has been previously described as being restricted to B and T lymphocytes, monocytes, macrophages, neutrophils, dendritic cells and some malignant cells derived from leukocytes [25,26]. CD37 expression on human B cells is able to associate noncovalently, with other TSPAN such as CD53, CD81, and CD82 [27]. Some authors suggest that CD37 could potentially be a therapeutic target for B- cell malignancies, with the use of small modular immunopharmaceuticals creating a novel class of therapies [28]. Other latter reports show that the CD37 gene is hypo methylated in mantle cell lymphoma, resulting in changes in mRNA levels. Hypo methylation of promoting regions of this gene, might be one reason for the lack or lower expression of this protein in our samples [29]. Although we did not perform hypo methylation analysis on any TSPAN in this study, investigation might be relevant. The results of CD81 and CD37 together, may play an important role in determining a good prognosis for sarcomas, since higher transcript expression levels were detected in lower grade or non-metastatic tumors.
Histological tumor type was also an important parameter, which we considered in these tumors and a comparative analysis of the TSPAN expression in all histological tumor types were performed. All synovial sarcomas and pleomorphic sarcomas in this study were of a high grade; although all the other tumor types comprised heterogeneous tumor grades. The expression of CD81 and CD37 mRNA were significantly higher in leiomyosarcomas when compared to synovial sarcomas. We hypothesized that this fact may be due to differences in the histological grade of the tumors, since all synovial sarcomas, samples present a high histological grade.
CD82, an important metastasis suppressor [5,7,30,31], showed a lower expression in synovial sarcomas compared to pleomorphic sarcomas. The same transcriptional expression profile was obtained with all the other TSPAN evaluated, although without any significant value. The differential profiles of TSPAN expression among synovial sarcomas and other histological types may be explained by differences in origin and cellular differentiation pathways, or by the inherently higher histological grade of synovial sarcomas.
CD9 and CD63, which have been widely discussed in literature, did not show any significant, statistical differences in transcript or protein expression levels within the analyzed groups. Molecules however, did present a trend of lower expression in our tumor samples, with poorer prognosis with marginal p value. Studies showed that metastatic melanoma lesions lack CD63 expression, but have a high expression in precursor lesions of melanoma [32,33]. Here, we found CD63 showing a slight trend associated with disease free survival. CD9 expression in tumors has demonstrated itself to be a good prognostic factor in breast, lung, colonic and pancreatic carcinoma [34,35]. Our group showed a CD9 downregulation associated with aggressive behavior in oral squamous cell carcinoma [18]. Another report has shown the CD9 differential expression in tumors is the result of changes to its 5´UTR region. These authors have found not only a reduction in CD9 mRNA expression, but also a distinct quantitative shift toward the long 5´ UTR in CD9 receptor negative cells [36]. This data allows us to state, that a lack of this protein might represent important alterations in cytoplasm pathways. Furthermore, an it could result in tumor aggressiveness, which brings this marker to a correlation with a good prognosis.
In the light of these results, TSPAN seems to present a defined pattern of expression in STS and their expression correlates with good prognosis. Particularly, CD37 and CD81, which show themselves to present higher expression in tumors with better prognosis (no metastatic and lower grade tumors). Thus, these two proteins may represent new prognostic markers in soft tissue sarcomas.
Acknowledgments
Financial support was provided by the CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and Fundação de Amparo à Pesquisa no Estado de São Paulo (FAPESP 98/14335-2). The authors thank to the Fundação Antônio Prudente, A. C. Camargo Cancer Center. We would like to thank José Ivanildo Neves, Carlos F. Nascimento and Severino S. Ferreira for their expert technical assistance.
References
- Skubitz KM, D'Adamo DR. Sarcoma. Mayo Clin Proc. 2007; 82: 1409-1432.
- Bains R, Magdum A, Bhat W, Roy A, Platt A, Stanley P. Soft tissue sarcoma - A review of presentation, management and outcomes in 110 patients. Surgeon. 2014.
- Karakousis CP, Emrich LJ, Rao U, Khalil M. Selective combination of modalities in soft tissue sarcomas: limb salvage and survival. Semin Surg Oncol. 1988; 4: 78-81.
- Thway K. Pathology of soft tissue sarcomas. Clin Oncol (R Coll Radiol). 2009; 21: 695-705.
- Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001; 58: 1189-1205.
- Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005; 6: 801-811.
- Lazo PA. Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 2007; 98: 1666-1677.
- Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014; 14: 49-60.
- Cajot JF, Sordat I, Silvestre T and Sordat B. Differential display cloning identifies motility related-protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res.1997; 57: 2593-2597.
- Jang HI, Lee H. A decrease in the expression of CD63 tetraspanin protein elevates invasive potential of human melanoma cells. Exp Mol Med. 2003; 35: 317-323.
- Kohno M, Hasegawa H, Miyake M, Yamamoto T, Fujita S. CD151 enhances cell motility and metastasis of cancer cells in the presence of focal adhesion kinase. Int J Cancer. 2002; 97: 336-343.
- Lee JH, Seo YW, Park SR, Kim YJ, Kim KK. Expression of a splice variant of KAI1, a tumor metastasis suppressor gene, influences tumor invasion and progression. Cancer Res. 2003; 63: 7247-7255.
- Cunha IW, Carvalho KC, Martins WK, Marques SM, Muto NH, Falzoni R, et al. Identification of genes associated with local aggressiveness and metastatic behavior in soft tissue tumors. Transl Oncol. 2010; 3: 23-32.
- Gomes LI, Silva RL, Stolf BS, Cristo EB, Hirata R, Soares FA, et al. Comparative analysis of amplified and nonamplified RNA for hybridization in cDNA microarray. Anal Biochem. 2003; 321: 244-251.
- Esteves GH, Simoes AC, Souza E, Dias RA, Ospina R, Venancio TM. New insights about host response to smallpox using microarray data. BMC Syst Biol. 2007; 1: 38.
- Lavorato-Rocha AM, de Melo Maia B, Rodrigues IS, Stiepcich MM, Baiocchi G, da Silva Cestari FM, et al. Prognostication of vulvar cancer based on p14ARF status: molecular assessment of transcript and protein. Ann Surg Oncol. 2013; 20: 31-39.
- Cunha IW, Lopes A, Falzoni R, Soares FA. Sarcomas often express constitutive nitric oxide synthases (NOS) but infrequently inducible NOS. Appl Immunohistochem Mol Morphol. 2006; 14: 404-410.
- Buim ME, Lourenço SV, Carvalho KC, Cardim R, Pereira C, Carvalho AL, et al. Downregulation of CD9 protein expression is associated with aggressive behavior of oral squamous cell carcinoma. Oral Oncol. 2010; 46: 166-171.
- Detchokul S, Williams ED, Parker MW, Frauman AG. Tetraspanins as regulators of the tumor microenvironment: implications for metastasis and therapeutic strategies. Br J Pharmacol. 2013; 171: 5462-5490.
- Sho M, Adachi M, Taki T, Hashida H, Konishi T, Huang CL, et al. Transmembrane 4 superfamily as a prognostic factor in pancreatic cancer. Int J Cancer. 1998; 79: 509-516.
- Hotta H, Ross AH, Huebner K, Isobe M, Wendeborn S, Chao MV, et al. Molecular cloning and characterization of an antigen associated with early stages of melanoma tumor progression. Cancer Res. 1988; 48: 2955-2962.
- Tohami T, Drucker L, Shapiro H, Radnay J, Lishner M. Overexpression of tetraspanins affects multiple myeloma cell survival and invasive potential. FASEB J. 2007; 21: 691-699.
- Luo RF, Zhao S, Tibshirani R, Myklebust JH, Sanyal M, Fernandez R, et al. CD81 protein is expressed at high levels in normal germinal center B cells and in subtypes of human lymphomas. Hum Pathol. 2010; 41: 271-280.
- Ma J, Liu R, Peng H, Zhou J, Li H. CD81 inhibits the proliferation of astrocytes by inducing G(0)/G (1) arrest in vitro. J Huazhong Univ Sci Technolog Med Sci. 2010; 30: 201-205.
- Charrin S, Jouannet S, Boucheix C, Rubinstein E. Tetraspanins at a glance. J Cell Sci. 2014; 127: 3641-3648.
- Lapalombella R, Yeh YY, Wang L, Ramanunni A, Rafiq S, Jha S, et al. Tetraspanin CD37 directly mediates transduction of survival and apoptotic signals. Cancer Cell. 2012; 21: 694-708.
- Angelisová P, Hilgert I, Horejsí V. Association of four antigens of the tetraspans family (CD37, CD53, TAPA-1, and R2/C33) with MHC class II glycoproteins. Immunogenetics. 1994; 39: 249-256.
- Zhao X, Lapalombella R, Joshi T, Cheney C, Gowda A, Hayden-Ledbetter MS, et al. Targeting CD37-positive lymphoid malignancies with a novel engineered small modular immunopharmaceutical. Blood. 2007; 110: 2569-2577.
- Leshchenko VV, Kuo PY, Shaknovich R, Yang DT, Gellen T, Petrich A, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010; 116: 1025-1034.
- Zhang XA, Lane WS, Charrin S, Rubinstein E, Liu L. EWI2/PGRL associates with the metastasis suppressor KAI1/CD82 and inhibits the migration of prostate cancer cells. Cancer Res. 2003; 63: 2665-2674.
- Malik FA, Sanders AJ, Jiang WG. KAI-1/CD82, the molecule and clinical implication in cancer and cancer metastasis. Histol Histopathol. 2009; 24: 519-530.
- Jang HI, Lee H. A decrease in the expression of CD63 tetraspanin protein elevates invasive potential of human melanoma cells. Exp Mol Med. 2003; 35: 317-323.
- Lupia A, Peppicelli S, Witort E, Bianchini F, Carloni V, Pimpinelli N, et al. CD63 tetraspanin is a negative driver of epithelial-to-mesenchymal transition in human melanoma cells. J Invest Dermatol. 2014; 134: 2947-2956.
- Miyake M, Nakano K, Itoi SI, Koh T, Taki T. Motility-related protein-1 (MRP-1/CD9) reduction as a factor of poor prognosis in breast cancer. Cancer Res. 1996; 56: 1244-1249.
- Xuan H, Hu X, Huang J. Role of motility-related protein-1 in promoting the development of several types of cancer (Review). Oncol Lett. 2014; 7: 611-615.
- Woegerbauer M, Thurnher D, Houben R, Pammer J, Kloimstein P, Heiduschka G, et al. Expression of the tetraspanins CD9, CD37, CD63, and CD151 in Merkel cell carcinoma: strong evidence for a posttranscriptional fine-tuning of CD9 gene expression. Mod Pathol. 2010; 23: 751-762.