
Research Article
J Dis Markers. 2020; 5(1): 1038.
Downregulation of Circulating Long Non-Coding Rnas GHRLOS and LINC00852 Associated with Type 2 Diabetes Mellitus
Al-Harithy R* and Anbari D
Department of Biochemistry, King AbdulAziz University, Saudi Arabia
*Corresponding author: Al-Harithy R, Biochemistry Department, King AbdulAziz University, Saudi Arabia
Received: September 08, 2020; Accepted: October 20, 2020; Published: October 27, 2020
Abstract
Long Non-Coding Rnas (lncRNAs) have an important role in many biological processes that are associated with several chronic diseases including Type 2 Diabetes Mellitus (T2DM). The aim of the present study is to identify novel biomarkers for T2DM by investigating the differentially expressed circulating lncRNAs, which are transcribed from Ghrelin (GHRL) Gene Region. Bioinformatic analysis was used to identify lncRNAs-GHRL candidates. The expression levels of the selected lncRNAs-GHRL were determined using Quantitative Real Time Polymerase Chain Reaction (qRT-PCR) assay on 62 diabetic patients and 32 non-diabetic controls. Receiver Operating Characteristic (ROC) curve was used to assess the discriminatory power of the candidate lncRNAs as biomarkers for T2DM. The expression profiles demonstrated that lncRNA GHRLOS and LINC00852 were significantly downregulated in the diabetic patients compared to the non-diabetic controls (P < 0.0001). The value of the Area Under The Curve (AUC) for GHRLOS and LINC00852 were 0.98 at a cut-off point of 1.03 and 0.96 at a cut-off value of 1.19; respectively. In conclusion, this study revealed that lncRNA GHRLOS and LINC00852 are novel biomarkers associated with T2DM and they might have regulatory roles in the development of the disease.
Keywords: Ghrelin (GHRL) gene; Long non-coding RNA; GHRLOS; LINC00852; Type 2 Diabetes Mellitus; Biomarker
Abbreviations
LncRNA: Long Non-Coding RNA; T2DM: Type 2 Diabetes Mellitus; GHRL: Ghrelin Gene; qRT-PCR: Quantitative Real Time Polymerase Chain Reaction; ROC: Receiver Operating Characteristic; GHRLOS: Ghrelin Gene Opposite Strand; AUC: Area Under The Curve; ADA : American Diabetes Association; HbA1c: Hemoglobin A1c; cDNA: Complementary DNA; GAPDH: Glyceraldehyde 3-Phosphate Dehydrogenase; BMI: Body Mass Index; NCD: Noncommunicable Diseases; MAPK: Mitogen-Activated Protein Kinase; S100A9:S100 CalciumBinding Protein A9; JNK: C-Jun N-Terminal Kinase; PI3K: Phosphatidylinositol 3-Kinase; Protein Kinase-Uncoupling Protein 2 (AMPK-UCP2)
Introduction
Type 2 Diabetes Mellitus (T2DM) is a complex metabolic disease characterized by insufficient insulin production and/or insulin resistance of human tissues [1,2]. It is a worldwide health crisis that leads to development of life-threatening health complications such as retinopathy, nephropathy, and neuropathy [3]. Despite of the great efforts that have been achieved in diagnosing and treating T2DM patients, the exact mechanism of its development remains poorly understood. Several investigations have been conducted to understand the pathogenesis of T2DM; however, the huge role that epigenetics may play in T2DM remains small compared to that devoted to traditional genetics work [4].
Epigenetic biomarkers have been identified and Long Noncoding Rnas (lncRNAs) are important regulators of the epigenetic status of the human genome. They are a type of Non-Coding Rnas (ncRNAs) which exceed 200 nucleotides in length without protein-coding potential [5,6]. The biogenesis process of lncRNAs is mainly similar to mRNA which is mediated by RNA polymerase II [7,8]. Moreover, different isoforms of lncRNAs can be transcribed from the same locus as a result of alternative splicing, cleavage and polyadenylation [9]. According to lncRNAs’ roles and biological positions, they can be classified as sense, antisense, intergenic and intronic lncRNAs [10]. The importance of lncRNAs are known in diverse of biological processes including cell’s functions, metabolism and genomic regulations [11,12]. The regulatory role may include the ability of lncRNAs to bind with Transcriptional Factors (TFs) and / or RNA polymerase II to transcription process. Also, they can indirectly regulate the expression level of mRNA by affecting its splicing, transporting and translation [5]. In addition, lncRNAs have been reported to contribute in genomic methylation and chromatin remodeling processes to regulate gene expression level [13]. Moreover, the presence of some genetic variation such as Single Nucleotide Polymorphisms (SNPs) within lncRNAs may contribute in modifying their secondary structure and altering their expression patterns. This would affect their regulatory role and consequently, contributing in the progression of the diseases [14]. To date, numerous studies have attempted to explain the association between lncRNAs and the development of various diseases [15,16]. However, the molecular mechanisms of lncRNAs in glucose homeostasis and T2DM remain elusive [17- 19]. The hunger hormone ghrelin or “lenomorelin” is a circulating peptide hormone which is secreted mainly from the stomach, while trace amounts derived from the pancreas [20,21]. Ghrelin is involved in several biological functions, which include body weight regulation, blood glucose homeostasis, appetite stimulation and insulin metabolism [22, 23]. Several studies have reported that insulin has inhibitory effect on ghrelin [23,24]. It has been suggested that ghrelin levels and insulin are inversely correlated in healthy human, which indicates the occurrence of feedback inhibition between them. Therefore, circulating ghrelin levels decreases in response of insulin secretion [25,26]. Previous studies revealed that ghrelin gene is strongly associated with insulin resistance and T2DM [27,28]. Thus, more Genetic and Epigenetic Studies on Ghrelin (GHRL) Gene are needed to understand its regulatory role in T2DM. The aim of this study is to identify potential circulating lncRNA-GHRL biomarkers that play a role in the development of T2DM.
Materials and Methods
Patients and Healthy Controls: This study was approved by the National Committee of Bio and Med. Ethics at King Abdul-Aziz University and Medical College, Jeddah, Saudi Arabia (reference number: HA-02-J-008). Between March 2019 and October 2019, patients with T2DM were recruited from the Diabetic and Endocrine Care Centre in Jeddah, Saudi Arabia. All participants provided informed consent prior to enrolment. All patients with T2DM were diagnosed according to the diagnostic criteria by the American Diabetes Association (ADA). All participants were Saudi, aged 40 years or older. For the diabetic patients, they were all diagnosed as diabetics since at least two years and they were taking diabetic medications such as metformin and insulin. For the non-diabetic controls, HbA1c level was between 4.8% and 5.6% and they were not under any medical conditions. Subject with independent chronic or acute medical conditions independent of diabetes and its complications were excluded from this study.
Blood Samples and Total RNA Extraction: Peripheral blood samples (5-8 ml) were obtained from each subject. The samples were collected in EDTA tubes. Total RNAs were isolated from whole blood samples using QIAamp RNA blood mini kit (Qiagen, USA) following the manufacturer’s instruction. RNase decontamination solution (RNaseZap) was used to prevent the degradation of RNA. The RNA concentrations were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Bioinformatics Analysis And Complementary DNA Synthesis(cDNA): LINCipedia database (https://lncipedia.org/) was used to determine lncRNAs that are related to GHRL gene. The analysis using LINCipedia database showed 11 GHRL-associated lncRNAs. For the cDNA synthesis, total RNA samples were reverse transcribed into cDNA using the total transcriptome cDNA synthesis kit (abm, Canada). All cDNA samples were diluted in 100 μl nucleasefree water and were stored at -20 °C prior to use.
Quantitative Real Time Polymerase Chain Reaction (Qrt-PCR) Analysis: Complementary Dna (cDNA) was amplified via qRTPCR using PowerUp Syber Green Master Mix kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Primers were purchased from Macrogen, Inc (Table 1). The thermocycling protocol included the following steps: initial incubation at 50ºC for 2 minutes, followed by 40 cycles amplification at 95ºC for 15 seconds, annealing at 55-60°C for 15 seconds, and extension at 72ºC for 1 minutes. Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) was used as an endogenous control to normalize the lncRNAs expression levels. All reactions were performed in triplicate.
Gene
Forward
Reverse
GAPDH
5'-TGTTCGTCATGGGTGTGAAC-3'
5'-ATGGCATGGACTGTGGTCAT-3'
GHRLOS
5'-TGGAAACTCCCCTAGCCACA-3'
5'-GCATCTCTCCTCTGTTCCGT-3'
LINC000852
5'-CGTTGCCTACAGTCAAGTCAGT-3'
5'-GCCATGGTTCCCTTACTGATAC-3'
Table 1: Primers used to validate gene expression.
Statistical Analysis: Three software were used for statistical analysis; Microsoft Excel, GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA), and MedCalc (MedCalc, Ostend, Belgium). The chi-square (χ2) test was used to compare distinct variables. The relative expression level of lncRNAs were calculated using 2−ΔΔCT method. Student’s t-test was used to compare the significance of gene expression levels between T2DM patients and non-diabetic controls. Unpaired t-test graph was generated to compare gene expression levels of lncRNAs and GHRL mRNA between the two groups. Pearson’s correlation coefficient (r) was calculated to determine the correlation of lncRNAs and GHRL mRNA to HbA1c and BMI. Receiver Operating Characteristic (ROC) curve was applied to define the discriminatory power of lncRNAs as biomarkers for T2DM. P-values (two-tailed) < 0.05 were considered statistically significant.
Results
Physical and Clinical Characteristics of Participants: The initial study cohort was 221 individuals, after applying the exclusion criteria only 62 diabetic patients (31 males and 31 females) and 32 non-diabetic controls (17 males and 15 females) were included in the study. No significant differences were noticed regarding gender (P = 0.77). There was a significant difference in age and BMI (P < 0.001). HbA1c was significantly (P < 0.001) higher in the diabetic patients compared to the non-diabetic controls.
Identification of Differentially Expressed Lncrnas: The expression pattern of the 11 candidate lncRNAs-GHRL and GHRL gene were measured by qRT-PCR using cDNA samples of 11 patients with T2DM and five non-diabetic controls. Out of the 11 LncRNAs, two lncRNAs, GHRLOS and LINC00852, were found to be differentially expressed in the T2DM patients compared to the non-diabetic controls. To validate the data, the total number of the cDNA samples (62 diabetic patients and 32 non-diabetic controls) were measured by qRT-PCR and showed that the expression level of lncRNA, GHRLOS and LINC00852, and GHRL mRNA were significantly (P < 0.0001) lower in the diabetic group with fold changes of 6.38 for GHRLOS, 8.02 for LINC00852, and 4.10 for GHRL mRNA (Figure 1).
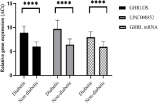
Figure 1: Differential expression of GHRLOS, LINC00852 and GHRL mRNA
between the diabetic patients and the non-diabetic controls.
Correlation of Lncrna GHRLOS and LINC00852 Expression with Hba1c and BMI: Pearson’s correlation coefficient (r) was used to evaluate the correlation between the expression levels of GHRLOS and LINC00852 and clinical characteristics. The results showed that the expression levels of GHRLOS and LINC00852 were negatively correlated with HbA1c (Table 2) and there is no correlation with BMI.
GHRLOS
LINC00852
Pearson’s correlation coefficient (r)
-0.54
-0.48
95% Confidence interval
0.383-0.672
0.313-0.626
P-value
<0.0001
<0.0001
Table 2: Correlation between HbA1c and lncRNAs.
Receiver Operating Characteristic (ROC) Curve Analysis of GHRLOS and LINC00852: To assess the discriminatory power of GHRLOS and LINC00852, ROC curve was performed and showed that both have high value of Area Under The ROC Curve (AUC) (Figure 2). The lncRNA GHRLOS and LINC00852 discriminated the T2DM patients from non-diabetic controls with high sensitivity and specificity (Table 3).
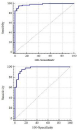
Figure 2: ROC curve of (A) GHRLOS and (B) LINC00852.
GHRLOS
LINC00852
Cut-off point
1.03
1.19
Sensitivity
Specificity
93.75
87.5
Youden index J
0.88
0.82
AUC
0.98
0.96
95% CI
0.930-0.998
0.911-0.994
Standard error
0.01
0.016
P-value
<0.0001
<0.0001
ROC: Receiver Operating Characteristic; AUC: Area Under the ROC Curve; CI: Confidence Interval; Youden index J: Defines the maximum potential effectiveness of a biomarker.
Table 3: ROC curve data.
Discussion
The World Health Organization’s (WHO) first target includes diabetes in its campaign for reducing premature death caused by Non-Communicable Diseases (NCD). T2DM, the most frequent subtype of diabetes, carries a 15% increase risk of premature death compare to healthy individuals [29]. In the last few years, there has been an increasing interest in studying the involvement of lncRNAs in the pathogenesis of different diseases including T2DM [30,31]. In this study, we identified two circulating lncRNAs related to GHRL gene, GHRLOS and LINC00852, that were downregulated in T2DM patients with high discriminatory power to distinguish between T2DM patients and non-diabetic individuals. This study opened a new avenue for exploring the use of GHRLOS and LINC00852 as biomarkers for early detection of T2DM.
Although the underlying causes that define the diabetic phenotype are extremely complicated, current view has valid the contributions of various lncRNAs as critical regulatory players linked to the progression of the disease [32,33]. Many relevant studies have indicated downregulation of lncRNAs expression in patients with T2DM versus control subjects, such as lncRNA ENST00000550337.1, THRIL, and SALRNA1 [34,35]. Our result match those observed in earlier studies and showed a significant downregulation in the expression level of lncRNA GHRLOS and LINC00852 in the peripheral blood of T2DM patients. Interestingly, Sathishkumar and his team stated that the expression levels of lncRNA THRIL and SALRNA1 in the diabetic patients were downregulated and were negatively correlated to glycemic control, insulin resistance, markers of senescence, and inflammation [35]. In another study, Wang et al., reported that dysregulated lncRNAs may have roles in T2DM pathogenesis through regulation of inflammation and insulin resistance [36]. Although these findings were important towards understanding the involvement of lncRNAs dysregulation during the development of T2DM, future studies are warranted to determine the precise regulatory mechanisms involved in the pathogenesis of T2DM. The findings from our study contribute towards the important role of lncRNA GHRLOS and LINC00852 in the pathophysiological processes of T2DM. Also, the correlations analysis between lncRNA GHRLOS and LINC00852 with HbA1c demonstrated an association between lncRNAs in peripheral whole blood and HbA1c level. Our findings suggest that lncRNAs, GHRLOS and LINC00852, may play a role in the regulation of glycometabolism.
It is well known that the major function of lncRNAs is to regulate the expression of protein-coding genes [37]. Intriguingly, our results showed a reduction in GHRL mRNA expression in T2DM patients when lncRNA GHRLOS and LINC00852 were decreased. The natural antisense transcript, GHRLOS, was first identified from the opposite strand of the GHRL gene by Seim and his colleagues and was found to transcribe natural antisense transcripts of GHRL gene [38]. A year later, the same group published an article about the complex organization of GHRLOS and concluded that the overlapping genomic arrangement of GHRLOS with the GHRL gene indicates that it is likely to have interesting regulatory and functional roles in the ghrelin axis [39]. In support to their suggestion, our data also implies that GHRLOS has a regulatory role in the expression of ghrelin levels.
In regard to LINC00852, transcript of GHRL, Liu and his team identified lncRNA LINC00852 as a potential target for the early prevention of spinal metastasis in lung adenocarcinoma. They also found that LINC00852 activates Mitogen-Activated Protein Kinase (MAPK) pathway by targeting S100 calcium-binding protein A9 (S100A9) that promote the progression of lung adenocarcinoma cells in vitro and in vivo. They also noticed that S100A9 has a strong activation effect on P38 and REK1/2 kinases and slight activation influence on the phosphorylation of the inflammatory C-Jun N-Terminal Kinase (JNK) in MAPK pathway [40]. Yung et al., found that JNK responds to hyperglycaemia and mediate the development of T2DM [41]. Furthermore, S100A9 has been known as a candidate gene for T2DM and may be involve in the pathogenesis of metabolic diseases [42-44]. It has been suggested that p38 MAPK, especially p38α MAPK, has a pathophysiological role in diabetes [45,46]. Moreover, it has been noticed the upregulation of p38 pathway in T2DM adipose tissue which might participate in the loss of Glucose Transporter Type 4 (GLUT4) expression [47]. Sweeney and his team proposed that phosphatidylinositol 3-kinase (PI3K) and p38 kinase stimulate glucose transport at cell membrane [48]. Considering all of these evidences, it can be concluded that LINC00852 may play a regulatory role in the progression of T2DM through MAPK family pathways and targeting S100A9.
It is well known that the reduction of insulin sensitivity in body’s tissues and cells is one of T2DM hallmark. According to previous study, insulin resistance is associated with (PI3K)/AKT pathway [49]. Moreover, a large body of researches contributed the fact that lncRNAs are key players in the process of insulin resistance as well as insulin signaling pathways [50-52]. The lncRNA H19 is a great example in which its ability to act as ‘sponge’ to regulate let-7 Micro Rnas (miRNAs) family in PI3K/AKT pathway[53]. Gao et al., study revealed that, in non-diabetic subjects, elevated level of H19 results a decrease in micro RNA let-7 level in PI3K/AKT pathway. Conversely, in hyperinsulinemia, PI3K/AKT pathway is activated and subsequently, the level of let-7 raises and H19 is rapidly depleted which can lead to insulin resistance and impairment of insulin signaling pathway. Their study indicated that H19 contributes in insulin resistance progression and in the development of T2DM through the regulation of (PI3K)/AKT pathway [54]. Interestingly, it has been established that GHRL has an inhibitory effect on insulin secretion via AMP‐Activated Protein Kinase (AMPK)–Uncoupling Protein 2 (UCP2) pathway [55] and also it regulate various biological processes in (PI3K)/AKT pathway [56-58]. Therefore, we propose that GHRLOS and LINC00852 may have similar activity to lncRNA H19 in the (PI3K)/AKT pathway and may inhibit insulin secretion through AMPK-UCP2 pathway as they both related to GHRL gene.
To the best of our knowledge, this study is the first to investigate the expression profiles of circulating lncRNAs, GHRLOS and LINC00852, in patients with T2DM and validate the utility of GHRLOS and LINC00852 as diagnostic biomarkers. The biomarkers identified in this study can be easily tested using peripheral blood with relatively low cost, high specificity, and sensitivity that make them potentially useful tools for the diagnosis of T2DM.
Conclusion
In summary, this research provides a framework for the exploration of differentially expressed circulating lncRNAs GHRLOS and LINC000852 in T2DM patients compared to non-diabetic individuals. It has also showed that those lncRNAs had significant discriminatory power to distinguish T2DM patients. Taken together, this study suggests that GHRLOS and LINC000852 may have regulatory roles in the development of T2DM and can perhaps be considered as promising biomarkers for early detection of T2DM.
Declaration of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
References
- Nolan CJ, Damm P, Prentki M. Type 2 diabetes across generations: from pathophysiology to prevention and management. The Lancet. 2011; 378: 169-81.
- Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel). 2015; 6: 87-123.
- Chawla A, Chawla R, Jaggi S. Microvasular and macrovascular complications in diabetes mellitus: Distinct or continuum? Indian J Endocrinol Metab. 2016; 20: 546-51.
- Bonnefond A, Froguel P. Rare and Common Genetic Events in Type 2 Diabetes: What Should Biologists Know? Cell Metab. 2015; 21: 357-68.
- Bhat SA, Ahmad SM, Mumtaz PT, Malik AA, Dar MA, Urwat U, et al. Long non-coding RNAs: Mechanism of action and functional utility. Non-coding RNA Research. 2016; 1: 43-50.
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009; 136: 629-41.
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nature Reviews Genetics. 2016; 17: 47-62.
- Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013; 9: e1003569.
- Ziegler C, Kretz M. The More the Merrier-Complexity in Long Non-Coding RNA Loci. Front Endocrinol (Lausanne). 2017; 8: 90.
- Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013; 10: 925-33.
- Li CJ, Xiao Y, Yang M, Su T, Sun X, Guo Q, et al. Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. Journal of Clinical Investigation. 2018; 128.
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nature Reviews Genetics. 2014; 15: 7-21.
- Zhao Y, Sun H, Wang H. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 2016; 6: 45.
- Li X, Wu Z, Fu X, Han W. lnc RNAs: insights into their function and mechanics in underlying disorders. Mutation research Reviews in mutation research. 2014; 762: 1-21.
- Jariwala N, Sarkar D. Emerging role of lnc RNA in cancer: a potential avenue in molecular medicine. Annals of translational medicine. 2016; 4: 286.
- Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lnc RNAs in the regulation of cancer stem cells. Cellular oncology (Dordrecht). 2018; 41: 585-603.
- Feng SD, Yang JH, Yao CH, Yang SS, Zhu ZM, Wu D, et al. Potential regulatory mechanisms of lncRNA in diabetes and its complications. Biochem Cell Biol. 2017; 95: 361-7.
- Sun X, Wong D. Long non-coding RNA-mediated regulation of glucose homeostasis and diabetes. Am J Cardiovasc Dis. 2016; 6: 17-25.
- Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nature Reviews Endocrinology. 2015; 11: 151-60.
- Date Y, Kojima M, Hosoda H, Sawaguchi A, Mondal MS, Suganuma T, et al. Ghrelin, a Novel Growth Hormone-Releasing Acylated Peptide, Is Synthesized in a Distinct Endocrine Cell Type in the Gastrointestinal Tracts of Rats and Humans.This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture, Endocrinology. 2000; 141: 4255-61.
- Broglio F, Gottero C, Benso A, Prodam F, Volante M, Destefanis S, et al. Ghrelin and the endocrine pancreas. Endocrine. 2003; 22:19-24.
- Arvat E, Di Vito L, Broglio F, Papotti M, Muccioli G, Dieguez C, et al. Preliminary evidence that Ghrelin, the natural GH secretagogue (GHS)- receptor ligand, strongly stimulates GH secretion in humans. Journal of Endocrinological Investigation. 2000; 23: 493-5.
- Möhlig M, Spranger J, Otto B, Ristow M, Tschöp M, Pfeiffer AFH. Euglycemic hyperinsulinemia, but not lipid infusion, decreases circulating ghrelin levels in humans. Journal of Endocrinological Investigation. 2002; 25: RC36-RC8.
- Saad MF, Bernaba B, Hwu CM, Jinagouda S, Fahmi S, Kogosov E, et al. Insulin Regulates Plasma Ghrelin Concentration. The Journal of Clinical Endocrinology & Metabolism. 2002; 87: 3997-4000.
- Cummings DE, Purnell JQ, Frayo RS, Schmidova K, Wisse BE, Weigle DS. A Preprandial Rise in Plasma Ghrelin Levels Suggests a Role in Meal Initiation in Humans. Diabetes. 2001; 50: 1714-9.
- Flanagan DE, Evans ML, Monsod TP, Rife F, Heptulla RA, Tamborlane WV, et al. The influence of insulin on circulating ghrelin. American Journal of Physiology-Endocrinology and Metabolism. 2003; 284: E313-E6.
- Al Qarni AA, Joatar FE, Das N, Awad M, Eltayeb M, Al-Zubair AG, et al. Association of Plasma Ghrelin Levels with Insulin Resistance in Type 2 Diabetes Mellitus among Saudi Subjects. Endocrinol Metab (Seoul). 2017; 32: 230-40.
- Dezaki K, Sone H, Yada T. Ghrelin is a physiological regulator of insulin release in pancreatic islets and glucose homeostasis. Pharmacology & therapeutics. 2008; 118: 239-49.
- Tancredi M, Rosengren A, Svensson AM, Kosiborod M, Pivodic A, Gudbjörnsdottir S, et al. Excess Mortality among Persons with Type 2 Diabetes. The New England journal of medicine. 2015; 373: 1720-32.
- He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. Lnc RNAs: key players and novel insights into diabetes mellitus. Oncotarget. 2017; 8: 71325-41.
- Ruan Y, Lin N, Ma Q, Chen R, Zhang Z, Wen W, et al. Circulating Lnc RNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function. Cellular Physiology and Biochemistry. 2018; 46: 335-50.
- Leti F, DiStefano JK. Long Noncoding RNAs as Diagnostic and Therapeutic Targets in Type 2 Diabetes and Related Complications. Genes (Basel). 2017; 8: 207.
- Goyal N, Kesharwani D, Datta M. Lnc-ing non-coding RNAs with metabolism and diabetes: roles of lncRNAs. Cellular and molecular life sciences : CMLS. 2018; 75: 1827-37.
- Li X, Zhao Z, Gao C, Rao L, Hao P, Jian D, et al. The Diagnostic Value of Whole Blood lnc RNA ENST00000550337.1 for Pre-Diabetes and Type 2 Diabetes Mellitus. Experimental and clinical endocrinology & diabetes : official journal, German Society of Endocrinology [and] German Diabetes Association. 2017; 125: 377-83.
- Sathishkumar C, Prabu P, Mohan V, Balasubramanyam M. Linking a role of lnc RNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018; 12: 41.
- Wang X, Chang X, Zhang P, Fan L, Zhou T, Sun K. Aberrant Expression of Long Non-Coding RNAs in Newly Diagnosed Type 2 Diabetes Indicates Potential Roles in Chronic Inflammation and Insulin Resistance. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2017; 43: 2367-78.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009; 10: 155-9.
- Seim I, Collet C, Herington AC, Chopin LK. Revised genomic structure of the human ghrelin gene and identification of novel exons, alternative splice variants and natural antisense transcripts. BMC genomics. 2007; 8: 298.
- Seim I, Carter SL, Herington AC, Chopin LK. Complex organisation and structure of the ghrelin antisense strand gene GHRLOS, a candidate noncoding RNA gene. BMC Mol Biol. 2008; 9: 95.
- Liu P, Wang H, Liang Y, Hu A, Xing R, Jiang L, et al. LINC00852 Promotes Lung Adenocarcinoma Spinal Metastasis by Targeting S100A9. Journal of Cancer. 2018; 9: 4139-49.
- Yung JHM, Giacca A. Role of c-Jun N-terminal Kinase (JNK) in Obesity and Type 2 Diabetes. Cells. 2020; 9: 706.
- Ortega FJ, Mercader JM, Moreno-Navarrete JM, Sabater M, Pueyo N, Valdés S, et al. Targeting the association of calgranulin B (S100A9) with insulin resistance and type 2 diabetes. Journal of Molecular Medicine. 2013; 91: 523-34.
- Catalán V, Gómez-Ambrosi J, Rodríguez A, Ramírez B, Rotellar F, Valentí V, et al. Increased levels of calprotectin in obesity are related to macrophage content: impact on inflammation and effect of weight loss. Molecular medicine (Cambridge, Mass). 2011; 17: 1157-67.
- Ortega FJ, Sabater M, Moreno-Navarrete JM, Pueyo N, Botas P, Delgado E, et al. Serum and urinary concentrations of calprotectin as markers of insulin resistance and type 2 diabetes. European journal of endocrinology. 2012; 167: 569-78.
- Westermann D, Rutschow S, Van Linthout S, Linderer A, Bücker-Gärtner C, Sobirey M, et al. Inhibition of p38 mitogen-activated protein kinase attenuates left ventricular dysfunction by mediating pro-inflammatory cardiac cytokine levels in a mouse model of diabetes mellitus. Diabetologia. 2006; 49: 2507- 13.
- Thandavarayan RA, Watanabe K, Ma M, Gurusamy N, Veeraveedu PT, Konishi T, et al. Dominant-negative p38alpha mitogen-activated protein kinase prevents cardiac apoptosis and remodeling after streptozotocininduced diabetes mellitus. American journal of physiology Heart and circulatory physiology. 2009; 297: H911-9.
- Carlson CJ, Koterski S, Sciotti RJ, Poccard GB, Rondinone CM. Enhanced Basal Activation of Mitogen-Activated Protein Kinases in Adipocytes From Type 2 Diabetes. Diabetes. 2003; 52: 634.
- Sweeney G, Somwar R, Ramlal T, Volchuk A, Ueyama A, Klip A. An inhibitor of p38 mitogen-activated protein kinase prevents insulin-stimulated glucose transport but not glucose transporter translocation in 3T3-L1 adipocytes and L6 myotubes. The Journal of biological chemistry. 1999; 274: 10071-8.
- Fernandez-Twinn DS, Alfaradhi MZ, Martin-Gronert MS, Duque-Guimaraes DE, Piekarz A, Ferland-McCollough D, et al. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol Metab. 2014; 3: 325-33.
- Degirmenci U, Li J, Lim YC, Siang DTC, Lin S, Liang H, et al. Silencing an insulin-induced lncRNA, Lnc ASIR, impairs the transcriptional response to insulin signalling in adipocytes. Scientific Reports. 2019; 9: 5608.
- Ellis BC, Graham LD, Molloy PL. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2014; 1843: 372-86.
- Chen B, Li J, Chi D, Sahnoune I, Calin S, Girnita L, et al. Non-Coding RNAs in IGF-1R Signaling Regulation: The Underlying Pathophysiological Link between Diabetes and Cancer. Cells. 2019; 8: 1638.
- Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lnc RNA antagonizes let-7 micro RNAs. Mol Cell. 2013; 52: 101-12.
- Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014; 42: 13799-811.
- Wang Y, Nishi M, Doi A, Shono T, Furukawa Y, Shimada T, et al. Ghrelin inhibits insulin secretion through the AMPK–UCP2 pathway in β cells. FEBS letters. 2010; 584: 1503-8.
- Lodeiro M, Theodoropoulou M, Pardo M, Casanueva FF, Camiña JP. c-Src regulates Akt signaling in response to ghrelin via beta-arrestin signalingindependent and dependent mechanisms. PLoS One. 2009; 4: e4686.
- Sun N, Wang H, Ma L, Lei P, Zhang Q. Ghrelin attenuates brain injury in septic mice via PI3K/Akt signaling activation. Brain Research Bulletin. 2016; 124: 278-85.
- Lien GS, Lin CH, Yang YL, Wu MS, Chen BC. Ghrelin induces colon cancer cell proliferation through the GHS-R, Ras, PI3K, Akt, and mTOR signaling pathways. European journal of pharmacology. 2016; 776: 124-31.