
Research Article
J Dis Markers. 2023; 8(1): 1050.
Immune Biomarker Combinations for Diagnosis Monitoring of Latent Tuberculosis Infection
Araujo Zaida2, Lopez-Ramos Juan Ernesto1, Enciso-Moreno Jose Antonio3, de Waard Jacobus Henri4, Rivas-Santiago Bruno5, Vanegas Magnolia6 and Patarroyo Manuel Alfonso6,7,8
1Laboratorio de Inmunologia de Enfermedades Infecciosas, Instituto de Biomedicina Dr, Jacinto Convit, Universidad Central de Venezuela, San Jose, Apartado 4043, Caracas 1010A, Caracas DC - Venezuela
2Centro de Estudios Cientiificos y Tecnologicos No. 18, Instituto Politecnico Nacional, Zacatecas, 98160 Zacatecas - Mexico
3Facultad de Química, Universidad Autónoma de Querétaro, México
4Laboratorio de Tuberculosis, Instituto de Biomedicina “Dr. Jacinto Convit”, Universidad Central de Venezuela, Venezuela
5Unidad de Investigación Biomédica de Zacatecas, Instituto Mexicano del Seguro Social, México
6Department of Molecular Biology and Immunology, Fundación Instituto de Inmunología de Colombia (FIDIC), Colombia
7Faculty of Medicine, Universidad Nacional de Colombia, Colombia
8Health Sciences Division, Main Campus, Universidad Santo Tomás, Colombia
*Corresponding author: Araujo Zaida Laboratorio de Inmunología de Enfermedades Infecciosas, Instituto de Biomedicina “Dr. Jacinto Convit”, Universidad Central de Venezuela, San José, Apartado 4043, Caracas 1010A, Caracas DC - Venezuela
Received: December 30, 2022; Accepted: February 03, 2023; Published: February 10, 2023
Abstract
Objective: Global Tuberculosis (TB) eradication efforts must also focus on detecting and treating cases of Latent TB Infection (LTBI); persons with LTBI can progress to active TB at any time, often many years or even decades after the initial infection, thereby serving as a source of new infections.
Methods: The aim was evaluated the diagnostic accuracy of the combination of serological host biomarkers that may support the differentiation between LTBI and Non-Infected (NI) individuals. A total of 182 adult Warao Amerindians were included; cases with LTBI (n=103) and Non-Infected (NI) individuals (n=79). The Real-time quantitative PCR (qPCR) was performed on all peripheral blood samples from Warao Amerindians and analyzed transcriptional immune biomarkers (i.e., IFN-γ, CD14, MMP-9, CCR5, CCL11, CXCL9/MIG, and uPAR/PLAUR proteins) under stimulation condition with ESAT-6, CFP10, and TB7.7 Mycobacterium Tuberculosis (Mtb)-antigens. Additionally, Enzyme-Linked Immunosorbent Assays (ELISA) were performed for evaluating host biomarker anti-synthetic peptides (5 ESAT-6 and 17 Ag85A synthetic peptides) covering Mtb antigen sequences.
Results: The approach’s diagnostic information was compared using Receiver Operating Characteristic (ROC) curves. The ROC analysis revealed high biosignature discriminative ability for the relative gene expression of MMP-9 high levels (AUC=0.799 ± 0.071: 0.640 - 0.917, 95% CI), p < 0.002) between LTBI and NI; additionally IgG anti-synthetic peptide; ESAT-6 P-12037 (AUC=0.640; 0.545-0.735 95% CI, p<0.007) allowed differentiation between LTBI and NI or healthy ones.
Conclusion: The accuracy of the MMP-9/IgG anti-P-12037 combination could have a high discriminative ability for diagnosing LTBI; such an approach holds promise for further validation.
Keywords: Tuberculosis (TB); Latent Tuberculosis Infection (LTBI); Warao Indigenous; Biomarker
Abbreviations: TB: Tuberculosis; ROC: Receiver Operating Characteristic; AUC: Area Under Curve; CI: Confidence Interval; WHO: World Health Organization; Mtb: Mycobacterium Tuberculosis; TST: Tuberculin Skin Test; ELISA: Enzyme-Linked ImmunoSorbent Assay; IgG: Immunoglobulin G; OD: Optical Density
Introduction
Tuberculosis (TB) is one of the top 10 causes of death worldwide [1]. The World Health Organization (WHO) report 2021 stated that reduced access to Active TB (ATB) diagnosis and treatment has resulted in an increase in TB deaths [1]. One third of the world’s population is latently infected with Mycobacterium Tuberculosis (Mtb) or Latent TB Infection (LTBI) and up to 10% of infected individuals develop active TB in their lifetime [2,3]. In Amerindian population (indigenous) tend to have much higher rates of TB in comparison with the general (non-indigenous) population [4]. Recently, it has been reported that in the admixed population of the Brazilian Amazon region; Amerindian ancestry in the 20–60% range was found to be the principal risk factor for increased susceptibility to TB [5].
Actually, the incidence of ATB by Mtb in Venezuela is increasing; [1] between 2014 and 2019 there was a rebound in cases (Creole or non-indigenous and indigenous individuals) that raised the rate to 39 per 100,000 inhabitants (39/100,000); the indigenous people (5.6%) being the most affected; so registering an abrupt increase in incidence in the last five years, going to exceed the levels reported three decades ago [1]. The National Integrated TB Control Program of Venezuela records the variables “indigenous” and “ethnicity” among the patient data; this has allowed the program to know the detailed statistics of the cases registered in indigenous patients. The Warao indigenous population is the second most important ethnic of the country with 48.771 inhabitants (6.7% of indigenous people), they mainly live in wooden houses raised on stilts along the Orinoco river’s banks (Delta’s rural areas). Data from the Coordination of the Regional Tuberculosis Control Program of the Delta Amacuro state reported that the incidence in the Warao population is 13 times higher than in the non-indigenous population and 19 times higher than the national rate [6-9]. Among the main factors that maintain the TB endemic are poverty, social inequality and inequity, migration, the impact of the HIV pandemic, and the lack of operational capacity for the detection of LTBI and ATB cases [10-12].
There are no diagnostic gold standards for LTBI, and all existing tests for LTBI are indirect approaches that provide immunological evidence of host sensitization to TB antigens [13,14]. Two tests currently used to diagnose LTBI, which are the Tuberculin Skin Test (TST) and the blood Interferon Gamma Release Assay (IGRA´s), both of which do not distinguish between ATB and LTBI. Positive response to IGRAs otherwise seem to confer only a small risk of reactivation (10–20 per 1000 person-years), which is similar to that of a positive TST [14]; however, the search for diagnostic and prognostic elements for LTBI continues. Clinical TB manifestations are closely related to the host immune response against Mtb. In individuals with different stages of Mtb infections, IgG antibody responses to Mtb antigens like ESAT-6 and Ag85 are frequently detected [15]. Additionally, biomarkers of innate and (Th1-) cell-mediated immunity are also observed [13,14]. Thus detection of multiple biomarkers non specific and specific for humoral as well as cellular immunity is necessary. Recently, we identified host biomarkers associated with LTBI in M. tuberculosis antigen-stimulated whole blood assays and IgG antibody serum from a TB endemic population in Delta’s Orinoco. A host biomarker signature of IgG anti-P-12037, MMP-9, CCR5 and CCL11 was identified; high IgG anti-P-12037 and MMP-9 and low CCR5 and CCL11 levels, relative to controls, were associated with LTBI [15,16].
Biomarkers are urgently needed to indicate progression from latent infection to clinical disease, to predict the risk of reactivation after cure, and to provide accurate end points for drug and vaccine trials [17-24]. Forty-eight analytes were evaluated by Luminex Assay in plasma obtained from whole blood stimulated cells. ROC curve analysis revealed that the combinations of 7 biomarkers resulted in the accurate prediction of 88.89% of ATB patients, 82.35% of subjects with LTBI and 90% of non-TB individuals [25]. These studies highlighted the importance of assessing multiple biomarkers for providing better understanding of protective biomarker profiles associated with resolution of clinical and subclinical infections in TB. The research proposal was evaluated the diagnostic accuracy of the combination of serological host biomarkers for identifying LTBI among Warao Amerindian communities, which would improve early diagnosis and allow a focus on detecting and treating cases of LTBI for improving TB control in Venezuela.
Methods
Study Population
A transversal study was carried out among individuals of both sexes from the Warao indigenous communities. A total of 182 adult Warao indigenous were included; cases with latent TB infection or LTBI (n=103) and non-infected (NI) individuals (n=79). The group of Warao indigenous in contact with active TB patients and the Tuberculin Skin Test (TST) positive, according to the WHO and the National Tuberculosis Control Program of Venezuela indigenous positive having ≥10 mm indurations were classified as LTBI; NI indigenous were also subjected to TST. Peripheral blood samples from both groups were drawn for QuantiFERON–TB Gold In-Tube (QFT-IT) testing (Commercial test QuantiFERON–TB Gold In-Tube, Cellestis, Victoria, Australia). Inclusion criteria took into account the recommendation previously reported [16].
Ethical Approval and Consent to Participate
This study was approved by the Institute of Biomedicine Central University of Venezuela Research Ethics Committee (protocol number FONACIT-2013002319/2013). All participating individuals signed voluntary informed consent forms.
Clinical Features, Microscopy, the Tuberculin Skin Test and Chest Radiograph
Individuals who had evidence of clinical symptoms suggesting pulmonary TB infection were excluded after having been diagnosed as having pulmonary TB using at least one of the following previously applied criteria published [16,26-28]. Individuals who had been prescribed immunosuppressive drugs (i.e., corticosteroids, azathioprine, and cyclophosphamide) were also excluded, as were participants who did not sign an informed consent agreement.
Isolated Blood Cells and Gene Biomarker Amplifications
Blood samples under non-stimulation conditions and stimulation conditions with M. tuberculosis antigens, Early Secretory Antigenic Target-6 (ESAT-6), Culture Filtrate Protein-10 (CFP10), and TB antigen 7.7 (TB7.7) were studied. A real-time reverse transcription polymerase chain reaction (RT-qPCR) assay was performed as previously reported [16].
Synthetic Peptides and Serological Assays
The solid-phase multiple peptide system was used for synthesising 22 peptides based on M. tuberculosis ESAT-6 and Ag85A amino acid (aa) sequences as previously reported [29,30]. The IgG levels against ESAT-6 and Ag85A peptides were determined in serum by ELISA assay as previously reported [15].
Statistical Analysis
Receiver Operating Characteristic (ROC) curves analysis, Student’s t-test, Fisher’s exact test, Mann Whitney test, Kruskal-Wallis and Dunn’s multiple comparison tests were performed as previously reported [15,16]. A scattergram was plotted using GraphPad Prism software version 5.02 (Trial version, GraphPadSofware, Inc., San Diego, USA). Statistically significant differences were those having a ≤0.05 p-value.
Results
The baseline characteristics of the participants are summarized in (Table 1). The TST+ status was significantly different between LTBI (78.0%) and NI (0.0%) groups, p< 0.0001; also for QuantiFERON–TB Gold In-Tube (QFT-IT) between LTBI (QFT-IT positive, 22.0%) and NI (QFT-IT negative, 0.0%) groups, p<0.001 (Table 1), and for CXRs, it was significantly different between LTBI (100.0%) and NI (0.0%) groups, p<0.0001 (Table 1).
Figure 1 shows the Optical Density (OD) distribution for the three best IgG anti-peptides between Warao indigenous LTBI and NI individuals; the IgG reactivity against the peptide is shown as values of the mean ± standard deviation (X ± SD). Mann Whitney test was used for comparing isotype reactivity differences between LTBI and NI groups. The LTBI’s IgG reactivity was high against 2 Ag85A peptides; P-10997 (0.538 ± 0.358) and P-11006 (0.493 ± 0.251) as compared with NI individuals (0.501 ± 0.327) (Figure 1B) and (0.461 ± 0.261) (Figure 1C), respectively; however, was no found significant difference. A significant difference was only found between LTBI (0.616 ± 0.244) and NI (0.512 ± 0.229) p<0.001 (Figure 1A) against M. tuberculosis ESAT-6 P-12037 amino acid (76-95 aa; ISEAGQAMASTEGNVTGMFA), (Figure 2), and ATB against M. tuberculosis ESAT-6 P-12035 amino acid (data not shown) (41-60 aa; AAWGGSGSEAYQGVQQKWDA), (Figure 2).
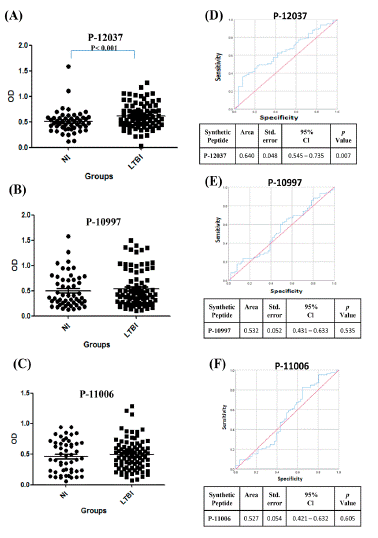
Figure 1: The IgG reactivity distribution against M. tuberculosis synthetic peptides and Receiver Operating Characteristic (ROC) curve analysis. The Optical density (OD) value distribution concerning antibody reactivities between Warao Amerindians with Latent Tuberculosis Infection (LTBI, ) and Non Infection (NI, ) or healthy individuals. OD of the anti IgG: P-12037 (A), P-10997 (B) and P-11006 (C). **P< 0.007.ROC curve analysis was used to evaluate the discriminating power of IgG anti-synthetic peptides between Latent Tuberculosis Infection (LTBI) and non‐infected (NI); P-12037 (D), P-10997 (E), P-11006 (F). Area Under the Curve (AUC), 95% confidence interval (95% CI) and p value< 0.05.
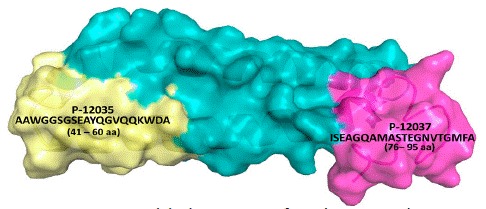
Figure 2: Amino acid (aa) sequence of synthetic peptide. Amino acid (aa) sequence of synthetic non-overlapping peptides spanning the sequence of ESAT-6 protein (P-12037 peptide). Protein Databank (PDB), (https://www.rcsb.org/structure/1WA8).
Marker
LTBI
NI
Age
40.0±12.91
37.79±13.44
Female (%)
54.6
58.8
Male (%)
45.3
41.2
TST+ (%)
78.0(a)
0.0(b)
QuantiFERONÒ (%)
Smear+ (%)22.0(c)
0.00.0(d)
0.0Culture+ (%)
Chest X-rays (CXRs) (%)0.0
100.0(e)n/d
0.0(f)
Table 1: Demographic and clinical information of the Warao indigenous enrolled in the study.
The approach’s diagnostic information was compared using Receiver Operating Characteristic (ROC) curves. The ROC analysis revealed good biosignature discriminative ability for serological biomarker IgG anti-P-12037 performed between LTBI and NI (AUC=0.640 ± 0.048: 0.545 - 0.735 95% CI), p<0.007 (Figure 1D); anti-P-12037 had 98.8% of sensitivity and 98.0% of specificity; the optimum cutoffs obtained for ESAT-6 P-12037 were; >0.171 and >1.428, respectively; while that the ROC analysis did not show good biosignature discriminative ability for serological biomarker for anti-P-10997 (Figure 1E), and anti-P-11006 (Figure 1F).
Figure 3 shows the approach’s diagnostic information for the relative gene expression of high (Figure 3A) and low (Figure 3B) levels of transcriptional serological immune biomarkers; such as interferon gamma (IFN-γ), the soluble phosphatidylinositol-linked membrane glycoprotein (sCD14), Matrix Metalloproteinases (MMP-9), CC receptor 5 (CCR5), and CC chemokine eotaxin (CCL11). ROC curve revealed good biosignature discriminative ability for the relative gene expression with high levels of MMP-9 performed between LTBI and NI (AUC=0.799 ± 0.071: 0.640-0.917, 95% CI), p<0.002), (Figure 3A); MMP-9 had 73.3% of sensitivity and 70.8% of specificity; the optimum cutoff obtained for MMP-9 was >0.880; the latter differentiated LTBI states from NI. Additionally, the relative gene expression low levels of CCL11 (AUC=0.277 ± 0.081: 0.020 - 0.119, 95% CI, p<0.02), (Figure 3A) and CCR5 (AUC=0.235 ± 0.072: 0.094 - 0.375, 95% CI, p<0.003), (Figure 3A) showed low sensitivity and specificity; while that the ROC analysis did not show adequate biosignature discriminative abilities for these serological biomarker (Figure 3A).
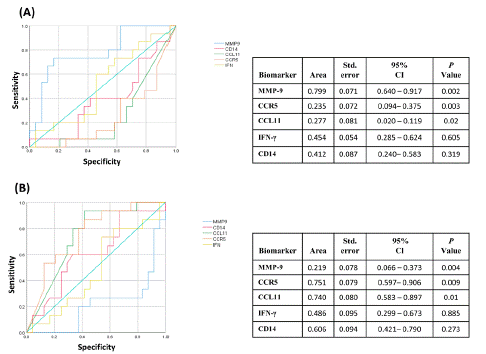
Figure 3: Receiver operating characteristic analysis of the mRNA expression levels. The ROC curve analysis was used to evaluate the discriminating power of serological host biomarkers between Latent Tuberculosis Infection (LTBI) and Non‐Infected (NI); A) The ROC data for considering the highest values as positive. B) The ROC data were obtained considering the smallest values as positive. Area Under the Curve (AUC), 95% confidence interval (95%CI) and p value < 0.05.
Considering revealed good biosignature discriminative ability for the relative gene expression with low levels of CCR5 performed between LTBI and NI (AUC=0.751 ± 0.079: 0.597 - 0.906, 95% CI), p<0.009) (Figure 3B), CCR5 had 80.0% of sensitivity and 62.5% of specificity; the optimum cutoff obtained for CCR5 was >0.667; low levels of CCR5 differentiated LTBI states from NI; additionally, the relative gene expression low levels of CCL11 (AUC=0.740 ± 0.080: 0.583 - 0.897, 95% CI, p<0.01) (Figure 3B), had 67.0% of sensitivity and 70.8% of specificity; the optimum cutoff obtained for CCRL11 was >0.500; low levels of CCL11 differentiated LTBI states from NI, and MMP-9 showed lower both sensitivity and specificity (AUC=0.219 ± 0.078: 0.066 - 0.373, 95% CI, p<0.004), (Figure 3B).
There were statistically significant differences; concerning TST+ status between (a) & (b) (p<0.0001), QuantiFERON–TB Gold In-Tube (c) & (d) (p<0.0001), and radiographical examinations performing Chest X-rays (CXRs) (e) & (f) (p<0.0001), n/d: test was not determined.
Discussion
Global TB eradication efforts must also focus on detecting and treating cases of LTBI [31,32]. At present, TST and IGRA tests are the most common tests for diagnosing LTBI. T-cell responses to Mtb like IGRAs are good diagnostic tools to define Mtb infection, which is the high-risk group in future TB reactivation. The use of biomarkers in patients with Mtb infection may further identify more specific populations who have current active TB disease or who will develop active TB in the future. For helping TB diagnosis, integrating biomarkers into clinical practice is suggested for a clinical TB suspect with negative preliminary workup and pending mycobacterial culture [33]. The present study evaluated the diagnostic accuracy of the combination of serological host biomarkers for identifying LTBI among Warao Amerindian communities. Taking into account that it had been reported current evidence on the contribution of B-cells and antibodies or Abs to immunity toward Mtb, their potential utility as biomarkers, and their functional contribution to Mtb control [34], we evaluated the diagnostic value and demonstrate the usefulness of serological methods studying the IgG or B-cell responses to ESAT-6 and Ag85A synthetic peptides. IgG against Mtb ESAT-6 P-12035 (amino acid 41-60 aa; AAWGGSGSEAYQGVQQKWDA) distinguished between Warao Amerindians with LTBI and ATB or individuals with active TB. However, the ROC analysis revealed good biosignature discriminative ability for the serological biomarker IgG anti-P-12037 performed between LTBI and NI; thus IgG Mtb ESAT-6 P-12037 (amino acid; 76-95 aa: ISEAGQAMASTEGNVTGMFA) could be used to distinguish between Warao Amerindians with LTBI and Non-Infected (NI) individuals. The antigen 85A (Ag85A) of Mtb for diagnostic purposes of LTBI among Warao Amerindians from a highly TB endemic area did not show discrimination between LTBI and NI [15]. The majority of Abs studies have been largely concentrated on their utility as diagnostic tools; with much attention for serological tests based on the detection of circulating Abs against Mtb specific antigens, because these have shown several advantages, as they are simple, cheap and feasible for point-of-care diagnostics [35,39].
Many attempts have been made to develop serologic LTBI and TB tests, which include the need to discriminate active TB or ATB from LTBI and finally able to perform consistently in genetically and immunologically diverse populations. In regard to the diagnostic accuracy of the MMP-9, CCR5, CCL11, CD14 and IFN- biomarkers detected by assessed in whole blood cultures using qPCR [40-43]. ROC analysis revealed good biosignature discriminative ability for the relative gene expression of MMP-9 high levels between LTBI and NI (AUC = 0.799 ± 0.071); MMP activity is an essential component of resistance to pulmonary mycobacterial infection and that MMP9, specifically is required for recruitment of macrophages and tissue remodeling to allow for the formation of tight, well-organized granulomas [16]. Combinations of immune biomarkers such as CXCL9, sCD14, MMP-9, and uPAR proteins, and anti- synthetic peptides, covering certain sequences of the ESAT-6 and Ag85A antigens of Mtb were evaluated among Venezuelan Creole or non-indigenous individuals; the findings showed that the IgG anti-P-12034/uPAR combination could be a potential biomarker for identifying clinical TB patients with 96.7% sensitivity [18].
As regard to CCR5, there was significantly statistically difference between the low CCR5 levels in the LTBI compared with the high CCR5 levels in the NI individuals. During Mtb infection, the pathogen modulates C-C Chemokine Receptor 5 (CCR5) to enhance IL-10 production, indicating the possible involvement of CCR5 in regulation of the host immune response; these events culminate in up regulation of the immunosuppressive cytokine IL-10 production, which is associated with the down-regulation of macrophage MHC-II expression along with the up-regulation of CCR5 expression via engagement of STAT-3 in a positive feedback loop [41]. The CCR5 immune biomarker has been reported as an immunological marker that differentiates between different outcomes of Mtb infection, CCR5 as well as CXCR4 have been related with the diagnosis of the extent of TB disease [42,43]. Additionally, the relative gene expression of CCL11 low levels differentiated LTBI state from NI or healthy ones. Eotaxin or CCL11 is a CC-chemokine that signals through the CCR3 receptor. It is produced by IFN-γ-stimulated endothelial cells and TNF-ɑ activated monocytes. It has been reported that CCL11, CCL24, and CCL26, which are produced by Th2 cells and other cells which induce Th2 development are increased in TB patients compared to controls; it seems that TB suppresses Th1 and the classic function of macrophages subsequently by inducing chemokines’ expression [44,45].
Authors have identified cytokines as host biosignature (IL-17F, MIP-3α, IL-13, IL-17A, IL-5, IL-9, IL-1β, IL-2 and IFN-γ) that could identify and uniquely discriminate between LTBI and ATB [46-48]. Present ROC analysis did not show good biosignature discriminative abilities for serological biomarker such as CD14 and IFN-γ. The IFN-γ biomarker was found to be expressed at low levels in the studied groups; the latter correlate with those of our previous study examining the capacity of antigen-induced proliferation by PBMCs and IFN-γ production in Warao indigenous with pulmonary TB and healthy controls. The IFN-γ production in Warao TB patients and controls was significantly lower after stimulation for 24 h and 48 h compared with that in the Creole group [43]. Biomarkers are urgently needed to indicate progression from latent infection to clinical disease, to predict the risk of reactivation after cure, and to provide accurate end points for drug and vaccine trials. Diagnosis and treatment of LTBI as well as ATB are required for effective TB control.
Conclusion
The ROC analysis revealed good biosignature discriminative ability for the MMP-9 and IgG anti-P-12037 indicating that both biomarkers are consistently associated with LTBI stage of M. tuberculosis infection. The relevance of the MMP-9 /IgG anti-P-12037 combination in a future point-of-care (POC) or a diagnostic POC test, which if validated, would contribute significantly to indicate the LTBI state, to predict the risk of reactivation after cure, to reduce the transmission, improving TB control in Venezuela.
Funding Sources
This work was supported by grants from the Central University of Venezuela [CDCH/UCV No. PG- 09-6256-8007/2011] and FONACIT [ No. 2013002319/2013].
Conflict of Interest
No conflict of interest to declare for all authors.
Acknowledgments
We are indebted to the indigenous Warao participants and non-participants in the study, to the translators of the Warao/Spanish language, to all of them for their human warmth and for having accepted us in their communities. Likewise, thanks are extended to the staff of the Delta Amacuro Regional Tuberculosis Control Program and the “Dr. Luis Razetti” from Tucupita, Delta Amacuro state.
Author Contributions
Z.A. took part in the study design, supervised the logistics work in the indigenous communities involved in data collection, and the assays developed at Laboratory. J.E.L.R. was involved in data analyses and data management. J.A.E.M. was involved in the analysis of molecular biomarkers, which were identified by genes overexpressed and associated with the development and progression of TB infections. J.H.de W. performed bacteriological and the tuberculin skin test and supervised the molecular assays for the confirmatory diagnosis of TB at Laboratory. B.R.S. was involved in data interpretation and critical revision of the manuscript. M.V. and M.A.P. performed synthetic peptides of sequences corresponding to the full length of both proteins: ESAT-6 and Ag85A.
References
- World Health Organization (WHO): Global Tuberculosis Report 2021, Geneva - Switzerland.
- Foulds J, O’Brien R. New tools for the diagnosis of tuberculosis: the perspective of developing countries. Int J Tuberc Lung Dis. 1998; 2: 778-783.
- Perkins MD. New diagnostic tools for tuberculosis. Int J Tuberc Lung Dis. 2000; 4: 182-188.
- Montenegro RA, Stephens C. Indigenous Health part 2: Indigenous health in Latin America and the Caribbean. Lancet. 2006; 367: 1859-−1869.
- Leal DFVB, Santana da Silva MN, Ricardo de Oliveira Fernandes DC, Gomes Rodrigues JC, da Costa Barros MC, do Carmo Pinto PD, et al. Amerindian genetic ancestry as a risk factor for tuberculosis in an amazonian population. PLoS One. 2020; 15: e0236033.
- Ministerio del Poder Popular para la Salud (MPPS). Programa Nacional Integrado de Control de la TB. Tuberculosis en población indígena Warao. Venezuela 2012 – 2017. Dirección General de Programas de Salud. 2018; Caracas – Venezuela.
- Instituto Nacional de Estadística (INE). Censo Nacional de Población y Vivienda 2011. Empadronamiento de la Población Indígena”. República Bolivariana de Venezuela, Ministerio del Poder Popular de Planificación, 2015; Caracas – Venezuela.
- Ministerio del Poder Popular para la Salud (MPPS). Anuario de morbilidad año 2013. MPPS-2015a; Caracas-Venezuela.
- Ministerio del Poder Popular para la Salud (MPPS). Anuario de mortalidad 2013. MPPS-2015b. Caracas-Venezuela.
- Narasimhan P, Wood J, Raina MacIntyre Ch, Mathai D. Risk Factors for Tuberculosis. Pulmonary Med. 2013; 2013: 828939.
- Tollefson D, Bloss E, Fanning A, Redd JT, Barker K, McCray E. Burden of tuberculosis in indigenous peoples globally: a systematic review. Int J Tuberc Lung Dis. 2013; 17: 1139-−1150.
- Malacarne J, Giraldo Rios DP, Furtado Passos da Silva CM, Braga JU, Bastos Camacho LA, Basta PC. Prevalence and factors associated with latent tuberculosis infection in a indigenous population in the Brazilian Amazon. Rev Soc Bras Med Trop. 2016; 49: 456-464.
- Barry CE 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, et al. The spectrum of latent tuberculosis: Rethinking the biology and intervention strategies. Nat Rev Microbiol. 2009; l7: 845-855.
- Pai M, Denkinger CM, Kik SV, Rangaka MX, Zwerling A, Oxlade O, et al. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev. 2014; 27: 3-20.
- Araujo Z, de Waard JH, Camargo M, Lopez-Ramos JE, Fernández de Larrea C, Vanegas M, et al. Diagnostic potential of the serological response to synthetic peptides from Mycobacterium tuberculosis antigens for discrimination between active and latent tuberculosis infections. Int J Pept Res Ther. 2022; 28: 1-12.
- Araujo Z, Palacios A, Enciso Moreno L, Lopez-Ramos JE, Wide A, de Waard JH, et al. Evaluation of the transcriptional immune biomarkers in peripheral blood from Warao indigenous associate with the infection by Mycobacterium tuberculosis. Rev. Soc. Brasileira Med. Trop. 2019; 52: e20180516.
- Walzl G, McNerney R, du Plessis N, Bates M, McHugh TD, Chegou, et al. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect Dis. 2018; 18: e199-e210.
- Araujo Z, Giampietro F, Bochichio MA, Palacios A, Dinis J, Isern J, et al. Immunologic evaluation and validation of methods using synthetic peptides derived from Mycobacterium tuberculosis for the diagnosis of tuberculosis infection. Mem Inst Oswaldo Cruz. 2013; 108: 131-139.
- Lopez Ramos JE, Macıas Segura N, Cuevas Cordoba B, Araujo Garcia Z, Bastian Y, Castaneda Delgado JE, et al. Improvement in the diagnosis of tuberculosis combining Mycobacterium tuberculosis immunodominant peptides and serum host biomarkers. Arch Med Res. 2018; 49: 147-153.
- Araujo Z, Macias Segura N, López Ramos JE, de Waard JH, Vanegas M, Patarroyo MA, et al. Diagnostic accuracy of combinations of serological biomarkers for identifying clinical tuberculosis. J Infect Dev Ctries. 2018; 12: 429-441.
- Wallis RS, Pai M, Menzies D, Doherty TM, Walzl G, Perkins MD, et al. Biomarkers and diagnostics for tuberculosis: progress, needs, and translation into practice. Lancet. 2010; 375: 1920-1937.
- Abramo C, Meijgaarden KE, Garcia D, Franken KLMC, Klein MR, Kolk AJ, et al. Monokine induced by interferon gamma and IFN-γ response to a fusion protein of Mycobacterium tuberculosis ESAT-6 and CFP-10 in Brazilian tuberculosis patients. Microbes Infect. 2006; 8: 45-51.
- Jenum S, Dhanasekaran S, Lodha R, Mukherjee A, Kumar Saini D, Singh S, et al. Approaching a diagnostic point-of-care test for pediatric tuberculosis through evaluation of immune biomarkers across the clinical disease spectrum. Sci. Rep. 2016; 6: 18520-18530.
- Chen Y, Wang J, Ge P, Cao D, Miao B, Robertson I, et al. Tissue inhibitor of metalloproteinases 1, a novel biomarker of tuberculosis. Mol Med. Rep. 2017; 15: 483-487.
- La Manna MP, Orlando V, Li Donni P, Sireci G, Di Carlo P, Cascio A, Dieli F, et al. Identification of plasma biomarkers for discrimination between tuberculosis infection/disease and pulmonary non tuberculosis disease. PLoS One. 2018; 13: e0192664.
- Kudoh S, Kudoh T. A simple technique for culturing tubercle bacilli. Bull. World Health Organ. 1974; 51: 71-82.
- Enciso Moreno JA, Monarrez Espino J. Performance of tuberculin skin test compared to QFT-IT to detect latent TB among high-risk contacts in Mexico. Arch Med Res. 2013; 44: 242-248.
- Arnadottir T, Rieder HI, Trebuq A, Waaler H. Guidelines for conducting tuberculin skin test surveys in high prevalence countries. Tuberc. Lung Dis. 1996; 77: 1-19.
- Tam JP, Heath WF, Merrifield RB. SN 1 and SN 2 mechanisms for the deprotection of synthetic peptides by hydrogen fluoride. Studies to minimize the tyrosine alkylation side reaction. Int J Pept Protein Res. 1983; 21: 57-65.
- Houghten RA. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc Natl Acad Sci USA. 1985; 82: 5131-5135.
- Lillebaek T, Dirksen A, Baess I, Strunge B, Thomsen VO, Andersen AB. Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. J Infect Dis. 2002; 185: 401-404.
- Dutta NK, Karakousis PC. Latent Tuberculosis Infection: Myths, models, and molecular mechanisms. Microbiol Mol Biol Rev. 2014; 78: 343.
- Chin-Chung S, Jann-Yuan W, Li-Na L, Chong-Jen Y, Kwen-Tay L. Improving tuberculosis diagnostics with biomarkers. Curr. Biomarker Findings. 2015; 5: 13-19.
- Rijink WF, Ottenhoff THM, Joosten SA. B-Cells and antibodies as contributors to effector immune responses in tuberculosis. Front Immunol. 2021; 12: e640168.
- Abebe F, Holm Hansen C, Wiker HG, Bjune G. Progress in serodiagnosis of Mycobacterium tuberculosis infection. Scand J Immunol. 2007; 66: 176-191.
- Ireton GC, Greenwald R, Liang H, Esfandiari J, Lyashchenko KP, Reed SG. Identification of Mycobacterium tuberculosis antigens of high serodiagnostic value. Clin Vaccine Immunol. 2010; 17: 1539-1547.
- Araujo Z, de Waard JH, Fernandez de Larrea C, López D, Fandiño C, Maldonado A, et al. Study of the Antibody Response against Mycobacterium tuberculosis Antigens in Warao Amerindian Children in Venezuela. Mem Inst Oswaldo Cruz. 2004; 99: 517-524.
- Araujo Z, Giampietro F, Castellano Cançado L, Singh M, Wide A. Comparison of serological responses in two different populations with pulmonary tuberculosis. Mem Inst Oswaldo Cruz. 2008; 103: 661-667.
- He XY, Li J, Hao J. Chen H-B, Zhao Y-Z Huang X-Y, et al. Assessment of five antigens from Mycobacterium tuberculosis for serodiagnosis of tuberculosis. Clin. Vaccine Immunol. 2011; 18: 565-570.
- Das S, Banerjee S, Majumdar S. Immune subversion by Mycobacterium tuberculosis through CCR5 mediated signaling: Involvement of IL-10. PLoS One. 2014; 9: e92477.
- Wolday D, Expression of chemokine receptors CCR5 and CXCR4 on CD4+ T cells and plasma chemokine levels during treatment of active tuberculosis in HIV-1-coinfected patients. J Acquir Immune Defic Syndr. 2005; 39: 265-271.
- Carpenter D, Taype C, Goulding J, Levin M, Eley B, Anderson S, et al. CCL3L1 copy number, CCR5 genotype and susceptibility to tuberculosis. BMC Med Genet. 2014; 15: 5-13.
- Sharifabadi RA, Hassanshahi G, Ghalebi SR, Arababadi MK, Khorramdelazad H, Zainodini N, et al. All eotaxins CCL11, CCL24 and CCL26 are increased but to various extents in pulmonary tuberculosis patients. Clin Lab. 2014; 60: 93-97.
- Giampietro F, de Waard JH, Rivas Santiago B, Enciso Moreno JA, Salgado A, Araujo Z. In vitro levels of cytokines in response to purified protein derivative (PPD) antigen in a population with high prevalence of pulmonary tuberculosis. Hum Immunol. 2010; 71: 1099-2104.
- Mihret A, Abebe M. Cytokines and Chemokines as Biomarkers of Tuberculosis. J Mycobac Dis. 2011; 3: 1000128.
- Yong YK, Tan HY, Saeidi A, Wong WF, Vignesh R, Velu V, et al. Immune Biomarkers for Diagnosis and Treatment Monitoring of Tuberculosis: Current Developments and Future Prospects. Front Microbiol. 2019; 10: 1-18.
- Chegou NN, Heyckendorf J, Walzl G, Lange Ch, Ruhwald M. Beyond the IFN-γ horizon: biomarkers for immunodiagnosis of infection with Mycobacterium tuberculosis. Eur Respir J. 2014; 43: 1472-1486.
- Kamakia R, Kiazyk S, Waruk J, Meyers A, Ochanda J, Ball TB, et al. Potential biomarkers associated with discrimination between latent and active pulmonary tuberculosis. Int J Tuberc Lung Dis. 2017; 21: 278-285.