
Research Article
J Dis Markers. 2023; 8(1): 1053.
Can Plasma miR-122-5p and miR-142-5p Predict Relapse in Metastatic Colorectal Cancer Patients?
Cervena K1,2*, Kubecek O3, Novosadova V4, Ryska P5, Petera J3 and Vymetalkova V1,2,6
1Institute of Experimental Medicine of Czech Academy of Sciences, Czech Republic
2Institute of Biology and Medical Genetics, 1st Medical Faculty, Charles University, Czech Republic
3Department of Oncology and Radiotherapy, Faculty of Medicine and University Hospital in Hradec Králové, Czech Republic
4Czech Centre for Phenogenomics, Institute of Molecular Genetics of the Czech Academy of Sciences, Czech Republic
5Department of Radiology, Faculty of Medicine and University Hospital in Hradec Kralove, Czech Republic
6Biomedical Centre, Faculty of Medicine in Pilsen, Charles University in Prague, Czech Republic
*Corresponding author: Cervena KDepartment of Molecular Biology of Cancer, Institute of Experimental Medicine of the Czech Academy of Sciences, Videnska 1083, 14200 Prague, Czech Republic
Received: January 24, 2023; Accepted: March 10, 2023; Published: March 17, 2023
Abstract
Approximately 20% of colorectal cancer patients already suffer from metastases at the time of diagnosis. Perhaps more serious, about 50% of the patients with localized tumours will eventually develop metastases during the course of their disease. A current hypothesis suggests that acquired or intrinsic chemoresistance is one of the major causes of metastasis. Recently, we found that circulating miR-122-5p and miR-142-5p could be used to predict both patient outcome and response to therapy. In the present study, we validate our previous results in patients with metastatic colorectal cancer (n=18), taking blood samples at three different times; i.e. metastasis resection surgery (T0), ~1 month after surgery (T1), and~8 months after surgery (T2). The expression levels of miR-122-5p and miR-142-5p were analysed in plasma and plasma extracellular vesicles (EVs) through RT-qPCR. The obtained results were compared against serum CEA and CA 19-9 levels, and computed tomography scans.
The rectal cancer patients in the good prognostic group showed significantly increased miR-142-5p expression profile between T1 and T2; whereas the poor prognostic group showed a lower expression during T2 when compared with T1. The decreased expression of miR-142-5p in the latter group could also be seen in the EVs of the patients. This is consistent with our previous study, where the downregulation of miR-142-5p could be associated with poor response to therapy. In addition, the individual analyses showed that the downregulation of both miRNAs predicted tumour relapse at an earlier time point than the markers CEA and CA 19-9.
In conclusion, the obtained data from this study demonstrates that miR-142-5p and miR-122-5p could be used as predictive bio-markers of tumour relapse in cases of metastatic rectal cancer, although further validation is still needed.
Keywords: Metastatic colorectal cancer; miR-122-5p; miR-142-5p; Relapse; Liquid biopsy
Abbreviations: CRC: Colorectal Cancer; miRNAs: MicroRNAs; RC: Rectal Cancer; mCRC: Metastatic Colorectal Cancer; EVs: Extracelullar Vesicles; RT-qPCR: Real-Time Quantitative Polymerase Chain Reaction; CT: Computed Tomography; LM: Liver Metastasis; DR: Disease Relapse
Introduction
Despite the improved methods of colorectal cancer (CRC) diagnosis and its increased therapeutic effectiveness, it still has a relatively high mortality rate [29]. Approximately 20% of CRC patients present metastatic disease at the time of diagnosis and about 50% of the patients with the localized disease will develop metastases during the course of therapy or soon thereafter [5]. The survival rate of these patients is ~90% for stage I; however, this becomes drastically lower (14%) in cases of metastatic CRC [30].
Acquired or intrinsic chemoresistance is a major cause of metastasis or recurrence in these patients, eventually leading to death [26,37]. Stage II and III patients present a recurrence rate of ~30-40%, becoming substantially higher for patients with metastatic disease [8,19]. Therefore, the identification of biomarkers predicting therapeutic response and relapse are crucial to prevent the early death of the patient, especially because only a few of them are currently used in clinical practice. The correct implementation of new biomarkers could facilitate patient stratification and personalized treatment [11].
We have previously reported that circulating microRNAs (miRNAs) - miR-122-5p and miR-142-5p - have great potential in the early detection of rectal cancer (RC) and prediction capacity for both outcome and response to therapy [4]. This study showed a different expression profile for both miRNAs when comparing RC patients and cancer-free individuals, the latter of which had significantly higher expression levels. Interestingly, the patients with good response showed increased miRNA expression after therapy whereas the unresponsive patients displayed consistently low expression levels of both miRNAs a year after diagnosis. MiRNAs can regulate the expression of target genes in a process known as RNA interference and many of them have essential roles in the preservation of life [20,22]. Further, miRNAs are relatively stable and can be found in body fluids, which enables their potential as cancer biomarkers [2].
In the present study, we validate the results obtained from our previous report through their application in metastatic colorectal cancer patients. Hereby we describe the expression profile of miR-122-5p and miR-142-5p in whole plasma and plasma extracellular vesicles (EVs) from metastatic colorectal cancer (mCRC) patients, making a simultaneous comparison with cancer-free individuals.
The expression profile of individual miRNA was compared against clinical data (i.e.tumour marker levels in serum, CT scans, and survival data) to determine their prognostic potential.
Material and Methods
Study Design
MiRNA expression levels were analysed through real-time quantitative polymerase chain reaction (RT-qPCR) in plasma and plasma EVs of mCRC patients (n=18). The samplings were taken at three different periods, i.e. metastasis resection surgery (T0), ~1 month after surgery (T1), and~8 months after surgery (T2). The obtained results were compared against a control group consisting of cancer-free individuals (n=51; Figure 1).
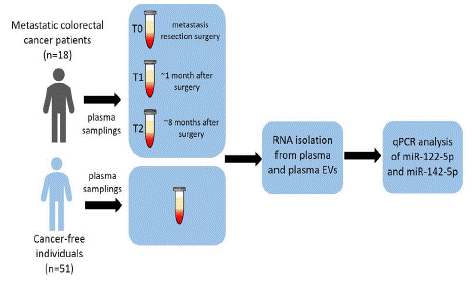
Figure 1: Schematic overview of the study.
RT-qPCR miRNA analysis (miR-122-5p and miR-142-5p) of mCRC patients (n=18) was performed on whole plasma and plasma EVs at 3 time periods: T0 (metastasis resection surgery), T1 (~1 month after surgery), and T2 (~8 months after surgery). The obtained results were compared against cancer-free individuals (n=51).
Study Population and Sample Collection
All the included individuals signed a written consent to participate in the study and approved the use of their biological samples for genetic analyses, all in accordance with the ethical principles described in the declaration of Helsinki. The study was approved by the Ethical Committee in the Faculty of Medicine and University Hospital in Hradec Kralove of Charles University and the Ethical Committee in the Institute of Experimental Medicine of the Czech Academy of Sciences.
The cohort comprised 18 histologically confirmed mCRC patients recruited from the Department of Oncology and Radiotherapy, University Hospital in Hradec Kralove, Czech Republic (2019 to 2021). The patients with no clinical history of concurrent malignancies, or CRC-associated well-defined inherited syndromes (e.g., Lynch syndrome, familial adenomatous, or MUTYH-associated polyposis) were included. Both plasma and clinical data from the included subjects were collected and supplemented with tumour marker levels in serum (i.e., CEA and CA 19-9) and CT scans (Table 1). The patients were under follow-up until August 2022. Two groups were made for this study, good prognostic group (i.e., those with no documented relapse following liver surgery) and the poor prognostic group (i.e., those with relapse following liver surgery). Clinical events and the therapeutic course are shown in Figure 2.
mCRC Patients
Cancer-Free Individuals
Characteristics
(n = 18)
Age (years)
Mean ± SD
64 ± 9
58 ±11
Gender
Male
5 (28%)
23 (45%)
Female
13 (72%)
28 (55%)
Tumour location
Coecum
1 (6%)
-
Ascending colon
1 (6%)
-
Splenic flexure
1 (6%)
-
Descending colon
1 (6%)
-
Sigmoid colon
7 (39%)
-
Rectosigmoid colon
1 (6%)
-
Rectum
6 (33%)
-
Tumour histology
Adenocarcinoma NOS
17 (94%)
-
Mucinous adenocarcinoma
1 (6%)
-
Tumour grade
1
0 (0%)
-
2
16 (89%)
-
3
2 (11%)
-
Time to liver metastasis1 (months)
Median (range)
18.5 (6.8–31.8)
-
Relapse-free survival2 (months)
Median (range)
15.1 (2.3–28.2)
-
Prognostic group
Good (no relapse following surgery)
6 (67%)
-
Poor (relapse following surgery)
12 (33%)
-
1Time from the diagnosis of the primary tumour to the detection time of liver metastasis
2Time from liver metastases resection to disease relapse
Table 1: Patient characteristics and survival data.
Figure 2: Swimmer plot.
Each bar represents a single subject in the study. The symbols along each bar represent various relevant clinical events.
Plasma Isolation
Peripheral blood was collected from all patients into EDTA tubes, and the plasma was separated through centrifugation at 1,400 rpm for 10 min/4°C within 1 hour of its collection. The plasma fraction was immediately stored at -80°C.
MiRNA Isolation
RNA was extracted from the plasma samples using a Plasma/Serum Circulating and Exosomal RNA Purification Kit (Norgen Biotek, Canada), according to the manufacturer´s directions. EVs were obtained by mixing 200 μl of the plasma samples with 50.4 μl Exo Quick exosome precipitation solution (System Biosciences, USA) and stored overnight at 4°C. The samples were then centrifuged at 1,500 g/4°C/30 min and the pellet dissolved in 200 μl of nuclease-free water. Total RNA was extracted immediately from the solution using Plasma/Serum Circulating and Exosomal RNA Purification Kit (Norgen Biotek, Canada) and quantified in a Qubit 3.0 Fluorometer using the Qubit microRNA assay Kit (Thermo Fisher Scientific, USA).
MiRNA Expression Analysis by RT-qPCR
The isolated RNA was reverse transcribed in a MJ Research PTC-200 Thermal Cycler (Marshall Scientific) using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, USA) according to the manufacturer’s directions. A Taqman microRNA assay (hsa-miR-142-5p - ID 002248, hsa-miR-122-5p – ID 002245, RNU48 – ID 001006, RNU6B – ID 001093, Applied Biosystems, and USA) was used for to evaluate the expression of the tested miRNAs. RNU48 and RNU6B were used as reference genes, as selected by Normfinder (Gen Ex Enterprise, MultiD, Thermo Fisher Scientific). The obtained cDNA was pre-amplified using an IQ SuperMix (Bio-Rad, USA), including 2 μl cDNA, 1.5 μl miRNA primer, 5 μl IQ SuperMix, and 1.5 μl RNAse free water, following the program 95°C/3 min, 95°C/15 sec - 59°C/4 min (18 cycles), and held at 4°C (MJ Research PTC-200 Thermal Cycler, Marshall Scientific). The pre-amplification samples were diluted 1:200. An Applied Biosystems 7500 Sequence Detection System was used for RT-qPCR. The 20 μl PCR reaction included 10 μl TaqMan Universal MasterMix II – no UNG (Applied Biosystems, USA), 1 μl Taqman microRNA assay, 7 μl RNAse-free water, and 2μl pre-amplified cDNA (200x dilution). The reactions were done in a 96-well optical plate at 95°C/10 min, followed by 95°C/15 sec - 60°C/10 min (40 cycles).
Statistical Analysis
The obtained data was captured in GenEx v6 (MultiD) and the statistical analyses were performed in the software R (v4.2.1) using a linear mixed model, where the random effect was set as ‘id’ and time as the variable of interest. Control samples were assigned an arbitrary time value (-1) because they were not measured repeatedly. Statistical significance was established as p≤0.05. Due to low number of samples data were analyzed independently for each condition.
Results
miR-122-5p and miR-142-5p Expression Analysis
Both miR-122-5p and miR-142-5p were successfully analyzed by RT-qPCR from the plasma and plasma EVs samples of mCRC patients (n=18) and cancer-free individuals (control group, n=51).
Plasma Levels
Compared with cancer-free individuals, plasma miR-142-5p expression levels of CRC patients were significantly downregulated during T2, showing a -2.62-fold change (p=0.005, Figure 3B). No significant difference was observed in the expression of miR-122-5p (Figure 3A). The patients were stratified according to tumour localization (colon and rectosigmoid vs. rectum) and according to the occurrence of relapse (further described as a good prognostic group or poor prognostic group). The patients with colon or rectosigmoid localized tumours showed significantly downregulated miR-142-5p expression (-3.01-fold change) during T2 in comparison with the cancer-free individuals (p=0.008, Figure 4). Interestingly, in patients with rectal cancer the expression of miR-142-5p was significantly different between the good and poor prognostic groups. Poor prognostic group showed a significant miR-142-5p upregulation (7.3-fold change) when compared with the good prognostic group in T1 (p=0.0094, Figure 5A). However, in T2 sampling the expression profiles were significantly modified: the miR-142-5p expression levels significantly increased in good prognostic group while in poor prognostic group during the same time period was observed decreased. Expression levels decreased when compared to second T1 sampling. This trend, which was associated with good response to therapy, was also observed in our previous study [4].
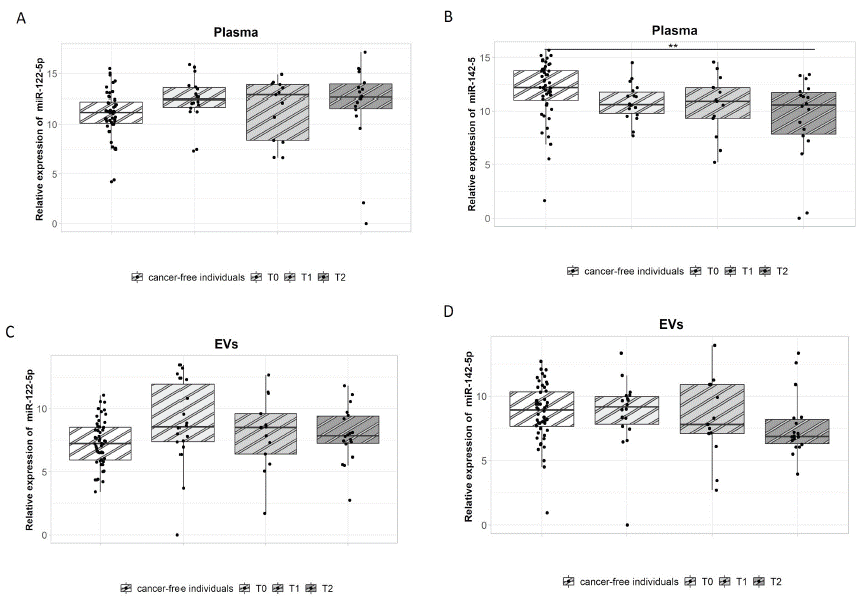
Figure 3: miR-122-5p and miR-142-5p expression analysis in plasma and plasma EVs.
(A,C) There was no significant difference in the expression of miR-122-5p between mCRC patients and cancer-free individuals. (B) Plasma miR-142-5p expression was significantly downregulated during T2 in mCRC patients in comparison with cancer-free individuals (p=0.005, -2.62-fold change).
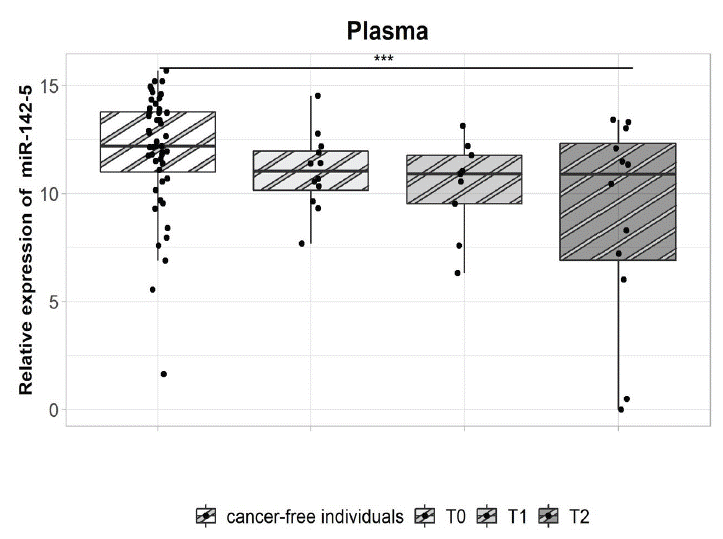
Figure 4: miR-142-5p expression analysis in the plasma of patients with metastatic colon and rectosigmoid cancer.
There is a significant difference in the expression of miR-142-5p during T2 in mCRC patients when compared to cancer-free individuals (p=0.008, -3.01-fold change).
EVs Levels
The analysis of plasma EVs revealed no differential miRNA expression between cancer-free individuals and mCRC patients. After stratifying the patients according to tumour localization, those with rectal cancer had lower miR-142-5p expression (3.8-fold change) in the poor prognostic group during T2 when compared to T1 (p=0.05, Figure 5B). This may suggest that the upregulation of miR-142-5p could be indicative of relapse.
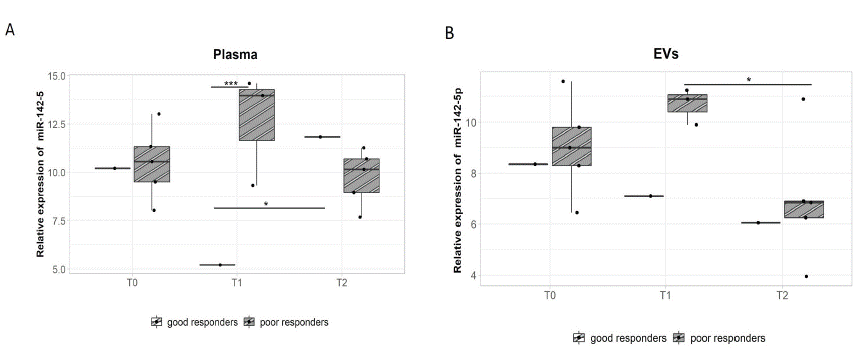
Figure 5: miR-142-5p expression analysis in the plasma and plasma EVs of patients with metastatic rectal cancer according to relapse.
A - The poor prognostic group had significantly higher miR-142-5p expression in plasma than the good prognostic group during T1 (p=0.0094, 7.3-fold change). However, there was a significant miR-142-5p up regulation during T2 in the good prognostic group (p=0.046, -6.6-fold change).
The Individual miRNA Expression and the Risk of Recurrence
In our study, we also focused on the individual miRNA expression levels. For a more detailed study, we present the follow-up data from two patients in the poor prognostic group and one from the good, including serum tumour markers CEA and CA 19-9, as well as CT scans.
Patient ID01, who was submitted to liver metastasis resection surgery, followed by fluoropyrimidine-based adjuvant chemotherapy for 3 months, showed decreasing expression levels of both miRNAs in plasma and plasma EVs after 8 months of the surgery and 12 months before a visible relapse could be observed in a CT scan. Noticeably, the level of the CEA and CA 19-9 tumour markers was not increased in serum at that time and no signs of liver metastases were observed on the CT scan (Figure 6).
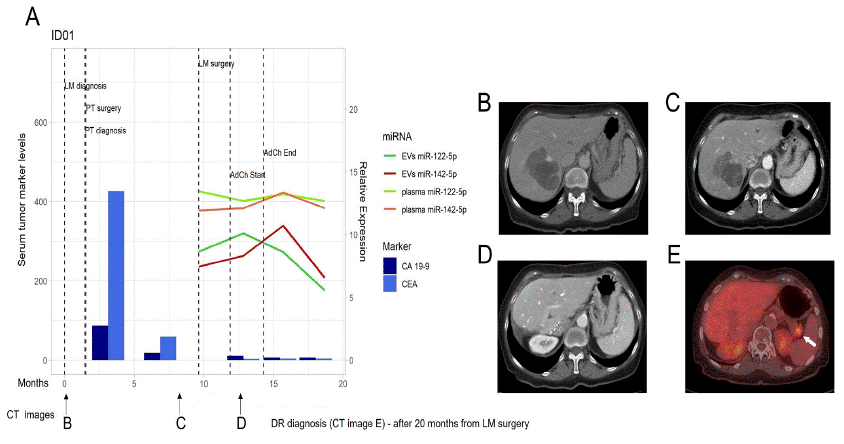
Figure 6: miR-122-5p and miR-142-5p expression in patient ID01.
A) miR-122-5p and miR-142-5p expression in plasma and plasma EVs in correlation with CA 19-9 and CEA in serum at the time of LM diagnosis, course of therapy, and follow up. Dark green line - EVs miR-122-5p, Red line – EVs miR-142-5p, Light green line – plasma miR-122-5p, Orange line – plasma miR-142-5p, Dark blue column – CA 19-9 levels, Light blue column – CEA levels. B) CT image at the time of LM diagnosis. C) CT image after Bevacizumab/mFOLFOX6 induction therapy (during which the patient had been diagnosed with stable disease according to RECIST criteria). D) CT image after LM resection and during the course of therapy. E) CT image after relapse (arrow). CT – Computed Tomography, EVs – Extracellular Vesicles, LM – Liver Metastasis, DR – Disease Relapse.
After liver metastasis resection surgery, and during the administration of adjuvant chemotherapy (mFOLFOX6/FUFA), patient ID03 displayed an increased expression of the tested miRNAs in both plasma and EVs (Figure 7). However, this upregulated expression rapidly declined after the end of therapy, with their baseline expression at T2, in both plasma and plasma EVs, being comparable to those observed prior to surgery. Further, a local liver relapse was detected after 5 months. This could imply that the downregulation of both miRNAs could predict these relapse events. It must be mentioned that the serum level of the tumour markers, CEA, and CA 19-9, remained unchanged during the entire follow-up period.
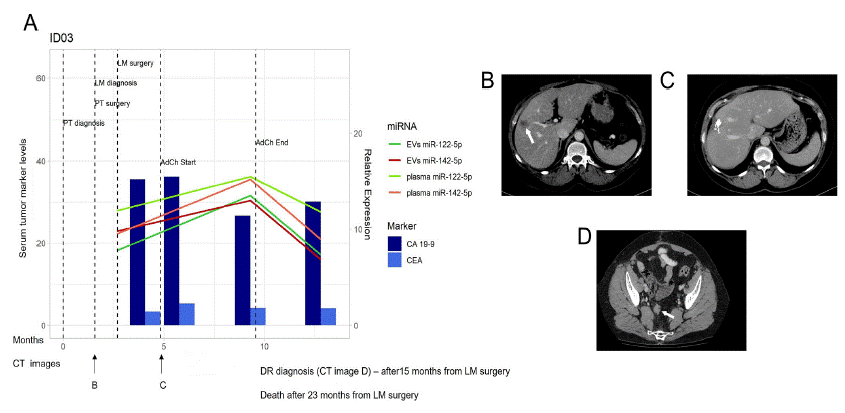
Figure 7: miR-122-5p and miR-142-5p expression in patient ID03.
A) miR-122-5p and miR-142-5p expression in plasma and EVs in correlation with CA 19-9 and CEA in serum at the time of LM diagnosis, course of therapy, and follow up. Dark green line - EVs miR-122-5p, Red line – EVs miR-142-5p, Light green line – plasma miR-122-5p, Orange line – plasma miR-142-5p, Dark blue column – CA 19-9 levels, Light blue column – CEA levels. B) CT image at the time of LM diagnosis (arrow). C) CT image after LM resection. D) CT image obtained during DR (arrow). CT – Computed Tomography, EVs – Extracellular Vesicles, LM – Liver Metastasis, DR – Disease Relapse.
Patient ID26, taken from the good prognosis group, did not develop relapse and the expression profile of both miRNAs, in either plasma or plasma EVs, was unstable during adjuvant chemotherapy (mFOLFOX6/FUFA, Figure 8). Regardless, a steady and progressive upregulation of these miRNAs was observed, in both plasma and EVs, after the end of therapy. This occurrence supports the hypothesis that a higher expression of these miRNAs is associated with a good response to therapy and, thereby, a good prognosis. Consistent with previous results, the levels of CEA and CA 19-9 in serum were unaffected during and after chemotherapy, remaining relatively stable since liver metastasis resection surgery.
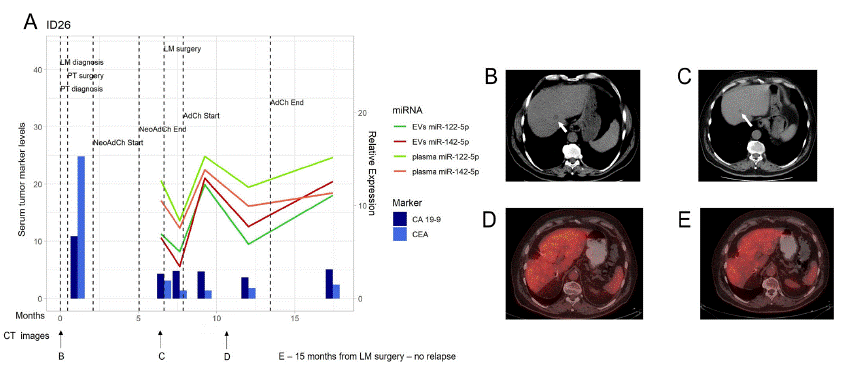
Figure 8: miR-122-5p and miR-142-5p expression in patient ID26.
A) miR-122-5p and miR-142-5p expression in plasma and EVs in correlation with CA 19-9 and CEA in serum at the time of LM diagnosis, course of therapy, and follow up. Dark green line - EVs miR-122-5p, Red line – EVs miR-142-5p, Light green line – plasma miR-122-5p, Orange line – plasma miR-142-5p, Dark blue column – CA 19-9 levels, Light blue column – CEA levels. B) CT image at the time of LM diagnosis (arrow). C) CT image after Bevacizumab/mFOLFOX6 induction therapy (during which the patient had been diagnosed with stable disease according to RECIST criteria). D) CT image after LM resection. E) CT image after 15 months of LM resection surgery, no sign of DR in liver. CT – Computed Tomography, EVs – Extracellular Vesicles, LM – Liver Metastasis, DR – Disease Relapse.
Discussion
Liver is the most common site for metastatic spread in CRC patients and liver metastasis is a leading cause of cancer mortality [18]. Currently, resection surgery is the only available treatment to improve their chance of survival. However, surgery is not possible in most cases and chemotherapy is thus recommended to relieve patients’ symptoms and prolong survival. Despite clinical efforts, chemoresistance and relapse are major obstacles during the course of therapy; therefore, there is an urgent need to discover and establish new biomarkers predicting the therapy response and outcome, as well as improve the chances of early detection of progression and/or relapse in mCRC patients. Several studies have focused on non-invasive approaches in recent years, e.g., liquid biopsy, that may enable early diagnosis and monitoring of the disease during treatment, enabling a more personalized therapy [23]. Ideally, the analysis of predictive biomarkers should be based on blood samples because of its availability. In this sense, miRNAs have been proven as a minimally invasive biomarker; however, this strategy is still in need of clinical validation.
In our previous study, we determined the expression profile of miR-122-5p and miR-142-5p in whole plasma and plasma EVs in rectal cancer patients [4]. The current study has expanded this analysis on mCRC patients exclusively. Previous results suggested that both miRNAs were significantly downregulated in rectal cancer patients when compared with cancer-free individuals. In the present study, this trend was also observed for miR-142-5p during T2. Interestingly, the poor prognostic group with mCRC had a significantly higher expression of miR-142-5p in plasma when compared to the good prognostic group after one month of LM resection surgery (T1). The good prognostic group with metastatic rectal cancer showed a significantly upregulated miR-142-5p expression between T1 and T2; however, its expression in the poor prognosis group decreased in comparison with T1. The same was also observed for miR-142-5p in the EVs from the poor prognostic group. This is consistent with our previous study, where the downregulation of miR-142-5p was also associated with poor response to therapy. Taking all into consideration, both studies strongly suggest that the level of miR-142-5p expression in plasma could be used as a predictive biomarker of relapse in patients with primary and metastatic rectal cancer. Although this trend was also observed for miR-122-5p in our previous study, the hereby shown results are inconsistent in comparison. This could be explained by the dynamic changes within the course of therapy. However, we did not have any samples prior to the commencement of therapy to make a more accurate assessment; further, the included mCRC patients in this study were subject to different therapeutical strategies (Figure 2). Chemo and radiotherapy can induce expression changes in several genes, altering the overall profile of a patient so that it could affect therapeutic response and, thus, their prognosis. It must be noted that the previous study consisted of rectal cancer patients only. This supports the claim that CRC is heterogeneous disease and colon, and rectum should be considered as separate diseases due to embryologic, anatomical and molecular differences including different therapy regimen and prognosis.
The identification of biomarkers for both therapeutic response and relapse would ease the implementation of personalized therapy by allowing the stratification of patients into appropriate treatment groups [41]. Therefore, we evaluated individual patients and compared the expression profile of the tested miRNAs with the values observed for tumor markers CEA and CA 19-9, and CT images obtained during the patient’s follow up. A similar trend was observed in this instance as well, i.e., the upregulation of both miRNAs was associated with good response to therapy and downregulation with poor response. Furthermore, in some cases, these miRNAs worked as better predictors of relapse than CEA or CA 19-9. These results suggest their immediate usefulness after the end of chemotherapy, as it is evident that there are huge dynamic changes in their expression [40].
Several CRC studies on miR-122-5p and miR-142-5p related to CRC have been previously discussed [4]. Thus, we focused on the correlation between the tested miRNAs with mCRC and liver metastasis. miR-122-5p has been previously associated with hepatocarcinogenesis, where its expression goes from non-tumor tissue to gradually decrease as chronic hepatitis, liver cirrhosis, primary hepatocellular carcinoma, and advanced carcinoma [34]. Because of its previous association with liver, miR-122-5p has been the subject of several studies on liver carcinoma, in which it shows an opposite trend to that observed in colorectal cancer, i.e.miR-122-5p is down regulated in tumor tissue when compared to non-tumor tissue [13,38]. The downregulation of miR-122-5p has also been associated with worse response to chemotherapy, shorter relapse-free survival, and metastasis-free survival [13,38]. The loss of miR-122-5p expression is known to increase tumor cell migration and invasion; therefore, miR-122-5p could be used as a marker of hepatocyte-specific differentiation [6,33].
The potential usefulness of other circulating miRNAs as biomarkers of mCRC has been extensively studied [1,12,24] MiR-203, miR-618, and miR-200c in serum were identified as promising biomarkers to predict metastasis [15,25,31,32]. These studies also showed the aberrant expression of miR-203 along the individual stages of carcinogenesis. Another study evaluated whether the combined analysis of CEA and miR-141 had greater sensitivity in the detection of mCRC [16]. The present study shows that, in some instances, CEA levels had lower sensitivity when detecting cases of recurrence when compared with the evaluated miRNAs. This was not unexpected, considering that as many as 50% of recurrence cases can be missed if only CEA is monitored during follow-up [28]. Thus, monitoring the expression of specific miRNAs could improve detection sensitivity during follow-up when compared to CEA alone. However, prospective studies are still needed to test the performance of individual miRNAs in terms of sensitivity and specificity. Besides plasma, some studies have also examined miRNA expression from plasma EVs. In this regard, the recurrence of liver metastasis has been linked with the expression of plasma exosomal miR-21 and miR-93-5p [7,35]. The dynamic expression of miRNAs during the administration of chemotherapy was also previously researched [17]. Schirripa et al., studied miRNAs expression profiles before and 15 days after therapy [27]. MiR-21, miR-141, miR-601, miR-221, and miR-760 were associated with the prediction of response to this therapy. An additional report revealed a higher miR-126 expression in non-responding patients, treated with bevacizumab for three weeks, when compared with patients with good response to the same therapy [14]. Other miRNAs, such as miR-20b-5p, miR-29b-3p, and miR-155-5p, can also be used to predict the response to bevacizumab therapy [36].
A growing number of ongoing and completed clinical trials showed miRNAs expand to preclinical and clinical research applications, although no cancer clinical trial has included miR-142-5p (clinicaltrials.gov). There are 13 clinical trials focusing on miR-122 but because of its specification, miR-122 is investigated in the liver in most clinical trials. In one of these trials, serum miR-122 was tested as a biomarker of drug-induced liver injury in patients with various malignant tumors (NCT03039062). There are ~10 clinical trials addressing the role of miRNAs in CRC, one of which is evaluating the impact of these miRNAs and their targets on the progression of CRC (NCT03309722). Authors have already identified highly expressed miR-153 in advanced CRC stages and this upregulation can affect cancer invasiveness and chemoresistance (i.e.,oxaliplatin and cisplatin) [39]. miR-224 and miR-19 have also been described as promoters of CRC metastasis through the downregulation of SMAD4 and TG2 [3,21]. Circulating miRNAs, found infaecal matter, have been included in a clinical trial in Spain (NCT05346757), which suggests that these miRNAs (including miR-421 and miR-27a-3p) could improve the detection sensitivity of advanced colorectal neoplasms in comparison to immunochemical tests [9,10]. Taking all these studies into consideration, it is clear that miRNAs can become a valuable tool in clinical applications.
The present study was limited by the size of the patient’s population, which included only 18 mCRC patients; further, we lacked either the primary or metastatic tumor tissue. On the other hand, the ability to perform repeated sample collections was a major advantage, i.e.T0 (at the time of metastasis resection surgery), T1 (~1 month after surgery), and T2 (~8 months after surgery). These samplings were enrolled in a single-center hospital and were processed by one competent person. Most mCRC studies do not include this type of repeated sample collection; therefore, they do not cover the dynamics of individual patients. In so doing, we obtained a wide spectrum of clinical information, which was further enriched by the analysis of conventional tumor markers (CEA, CA-19-9) and CT scan images. Moreover, plasma is a great source that brings advantages of the easy and non-invasive collection over time.
Despite the above-mentioned limitations of the study, we can suggest with a fair degree of certainty that the upregulation of miR-142-5 in plasma can predict relapse events in patients with primary and metastatic rectal cancer. On the other hand, the expression of miR-122 was not different in mCRC patients but in individual monitoring, the downregulation of both miRNAs in plasma and plasma EVs predicted events of relapse earlier than the already established tumor markers. In conclusion, the obtained results suggest that miR-142-5p and miR-122-5p can be used as predictors of relapse in primary and metastatic rectal cancer patients; however, further analysis, including the molecular stratification and localization of patients, is still needed to fully validate our observations.
Acknowledgements
This project was supported by the Czech health research council of the Ministry of Health of the Czech Republic (NV19–09-00237). We are thankful to the Cooperatio Program, research area “Oncology and Haematology”. Thanks to Dr. Daniel Díaz for his kind assistance in proofreading and editing this manuscript.
Author Contributions
Study design and execution – VV and KC. Collection and preparation of samples – KC, OK, JP and PR. Statistical analysis – VN. Interpretation of data – VV, KC, and OK. Original draft preparation – KC, VV, and OK. Writing – reviewing and editing – VV, KC, VN, OK and JP. All authors contributed to the article and approved the submitted version.
Clinical Trials
Binghe X., Qiao L. Serum miR-122 as a Real-time Detection Biomarker of Drug-induced Liver injury by Chemotherapy. ClinicalTrials.gov. NCT03039062.
Mirnezami A., Peppa N. Molecular Pathology of Colorectal Cancer: Investigating the role of Novel Molecular Profiles, microRNA´s, and their Targets in Colorectal Cancer Progression. ClinicalTrials.gov. NCT03309722.
Diaz M., Castells A. Validation of a microRNA based Fecal (miRFec) test for Colorectal Cancer screening (miRFec). ClinicalTrials.gov. NCT05346757.
References
- Balacescu O, Sur D, Cainap C, Visan S, Cruceriu D, et al. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int J Mol Sci. 2018; 19: 3711.
- Buhagiar A, Seria E, Borg M, Borg J, Ayers D. Overview of microRNAs as liquid biopsy biomarkers for colorectal cancer sub-type profiling and chemoresistance. Cancer Drug Resist. 2021; 4: 934-945.
- Cellura D, Pickard K, Quaratino S, Parker H, Strefford JC, et al. miR-19-Mediated Inhibition of Transglutaminase-2 Leads to Enhanced Invasion and Metastasis in Colorectal Cancer. Mol Cancer Res. 2015; 13: 1095-1105.
- Cervena K, Novosadova V, Pardini B, Naccarati A, Oppattova A, et al. Analysis of MicroRNA Expression Changes During the Course of Therapy In Rectal Cancer Patients. Front Oncol. 2021; 11: 702258.
- Ciardiello F, Ciardiello D, Martini G, Napolitano S, Tabernero J, et al. Clinical management of metastatic colorectal cancer in the era of precision medi-cine. CA Cancer J Clin. 022; 72: 372-401.
- Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009; 28: 3526-36.
- Despotovic J, Bogdanovic A, Dragicevic S, Galun D, Krivokapic Z, et al. Prognostic potential of circulating miR-93-5p in patients with colorectal cancer liver metastases. Neoplasma. 2022; 69: 430-442.
- Duineveld LA, van Asselt KM, Bemelman WA, Smits AB, Tanis PJ, et al. Symptomatic and Asymptomatic Colon Cancer Recurrence: A Multicenter Cohort Study. Ann Fam Med. 2016; 14: 215-20.
- Duran-Sanchon S, Moreno L, Auge JM, Serra-Burriel M, Cuatrecasas M, et al. Identification and Validation of MicroRNA Profiles in Fecal Samples for Detection of Colorectal Cancer. Gastroenterology. 2020; 158: 947-957 e4.
- Duran-Sanchon S, Moreno L, Gomez-Matas J, Auge JM, Serra-Burriel M, et al. Fecal MicroRNA-Based Algorithm Increases Effectiveness of Fecal Immunochemical Test-Based Screening for Colorectal Cancer. Clin Gastroenterol Hepatol. 2021; 19: 323-330 e1.
- Filip S, Vymetalkova V, Petera J, Vodickova L, Kubecek O, et al. Distant Metastasis in Colorectal Cancer Patients-Do We Have New Predicting Clinicopathological and Molecular Biomarkers? A Comprehensive Review. Int J Mol Sci. 2020; 21: 5255.
- Gherman A, Balacescu L, Gheorghe-Cetean S, Vlad C, Balacescu O, et al. Current and New Predictors for Treatment Response in Metastatic Colorectal Can-cer. The Role of Circulating miRNAs as Biomarkers. Int J Mol Sci. 2020; 21: 2089.
- Ha SY, Yu JI, Choi C, Kang SY, Joh JW, et al. Prognostic significance of miR-122 expression after curative resection in patients with hepatocellular carcinoma. Sci Rep. 2019; 9: 14738.
- Hansen TF, Carlsen Al, Heegaard NHH, Sorensen FB, Jakobsen A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer. 2015;112: 624-9.
- Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017; 66: 654-665.
- Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011; 6: e17745.
- Kjersem JB, Ikdahl T, Lingjaerde OC, Guren T, Tveit KM, et al. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer pa-tients receiving first-line oxaliplatin-based treatment. Mol Oncol. 2014; 8: 59-67.
- Kow AWC. Hepatic metastasis from colorectal cancer. J Gastrointest Oncol. 2019; 10: 1274-1298.
- Kunst N, Escudero FA, Aas E, Coupe VMH, Schrag D, et al. Estimating Population-Based Recurrence Rates of Colorectal Cancer over Time in the United States. Cancer Epidemiol Biomarkers Prev. 2020; 29: 2710-2718.
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense com-plementarity to lin-14. Cell. 1993; 75: 843-54.
- Ling H, Pickard K, Ivan C, Isella C, Ikuo M, et al. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut. 2016; 65: 977-989.
- Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015; 34: 5003-11.
- Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022; 21: 79.
- Nassar FJ, Msheik ZS, Itani MM, Helou RE, Hadla R, et al. Circulating miRNA as Biomarkers for Colorectal Cancer Diagnosis and Liver Metasta-sis. Diagnostics (Basel). 2021; 11: 341.
- Radanova M, Mihaylova G, Mihaylova Z, Ivanova D, Tasinov O, et al. Circulating miR-618 Has Prognostic Significance in Patients with Metastatic Colon Cancer. Curr Oncol. 2021; 28: 1204-1215.
- Ramos A, Sadeghi S, Tabatabaeian H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int J Mol Sci. 2021; 22: 9451.
- Schirripa M, Borelli B, D’AurizoR, Lubrano S, Cremolini C, et al. Early modifications of circulating microRNAs levels in metastatic colorectal cancer patients treated with regorafenib. Pharmacogenomics J. 2019; 19: 455-464.
- Shinkins B, et al. The diagnostic accuracy of a single CEA blood test in detecting colorectal cancer recurrence: Results from the FACS trial. PLoS One. 2017; 12: e0171810.
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022; 72: 7-33.
- Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020; 70: 145-164.
- Takano Y, Masuda T, Inuma H, Yamaguchi R, Sato K, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via induc-ing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017; 8: 78598-78613.
- Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, et al. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in pa-tients with colorectal cancer. Ann Surg. 2014; 259: 735-43.
- Tsai WC, Hsu PWC, Lai TC, Chau GY, Lin CW, et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009; 49: 1571-82.
- Tsang FH, Au SLK, Wei L, Fan DNY, Lee JMF, et al. MicroRNA-142-3p and microRNA-142-5p are downregulated in hepatocellular car-cinoma and exhibit synergistic effects on cell motility. Front Med. 2015; 9: 331-43.
- Tsukamoto M, Linuma H, Yagi T, Matsuda K, Hashiguchi Y, et al. Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology. 2017; 92: 360-370.
- Ulivi P, Canale M, Passardi A, Marisi G, Valgiusti M, et al. Circulating Plasma Levels of miR-20b, miR-29b and miR-155 as Predictors of Bevaci-zumab Efficacy in Patients with Metastatic Colorectal Cancer. Int J Mol Sci. 2018; 19: 307.
- Vodenkova S, Buchler T, Cervena K, Veskenova V, Vodicka P, et al. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol Ther. 2020; 206: 107447.
- Zhan G, Jiang H, Yang R, Yang K, miR-122 and miR-197 expressions in hepatic carcinoma patients before and after chemotherapy and their effect on patient prognosis. Am J Transl Res. 2021; 13: 6731-6737.
- Zhang L, Pickard K, Jenei V, Bullock MD, Bruce A, et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013; 73: 6435-47.
- Zhang N, Hu X, Du Y, Du J. The role of miRNAs in colorectal cancer progression and chemoradiother-apy. Biomed Pharmacother. 2021; 134: 111099.
- Zhou Q, Perakis SO, Ulz P, Mohan S, Riedl JM, et al. Cell-free DNA analysis reveals POLR1D-mediated resistance to bevacizumab in colorectal cancer. Genome Med. 2020; 12: 20.