
Special Article - Antisense Drug Research and Development
J Drug Discov Develop and Deliv. 2016; 3(1): 1020.
Custom Cocktail for Curing any Cancer: A Strategy for Destroying any Cancer without Harming the Patient
Summerton J*
Gene Tools, LLC, USA
*Corresponding author: Summerton J, Gene Tools, LLC, 1001 Summerton Way, Philomath, Oregon 97370, USA
Received: January 28, 2016; Accepted: April 27, 2016; Published: May 02, 2016
Abstract
Drugs for curing any cancer must completely destroy a patient’s cancer without harming the patient. To avoid harming the patient, precision-targeted drugs are used to inactivate RNA transcripts that are absent from the patient’s normal cells, but present in and essential to the viability of the patient’s cancer. Such “cancer-essential” transcripts are from embryo-active genes normally deactivated in adults, but reactivated in cancers, or from oncogenic viruses. For complete destruction of the patient’s cancer a “custom cocktail” of precisiontargeted drugs will target multiple cancer-essential transcripts found to be present in and essential to that patient’s cancer.
This paper describes: the basis for selecting targets in a patient’s cancer; selecting drugs for precisely inactivating those targets; and, assembly of a custom cocktail of such drugs designed to provide a safe and effective cure for that patient’s cancer. Preparation and use of a custom cocktail for a specific patient entails:
1. A biopsy of the patient’s cancer is sequenced to identify the canceressential transcripts present in that cancer.
2. With that information, assemble a cocktail of precision-targeted drugs effective against multiple cancer-essential transcripts found to be present in that patient’s cancer, and treat the patient with that custom cocktail.
Also described is a basis for possibly obtaining prompt and affordable regulatory approvals for such cocktails.
I believe this therapeutics strategy solves the central challenge in treating cancers: how to completely destroy any cancer without harming the patient. It also offers a direct and rapid route to a safe, effective, and affordable treatment for any cancer.
Keywords: Custom cocktails; Blocking cancer-essential transcripts
Introduction
Ideal cures for cancers should precisely attack and completely destroy any patient’s cancer - without damage to the patient. And such cures should be affordable. But two technical challenges have long foiled success in the quest for such ideal cures:
1. Cancer cells evolve from the patient’s normal cells and so both cell types share nearly all of the same genes, gene transcripts, and proteins. Thus it is very difficult to completely destroy the cancer cells without also damaging or killing the patient.
2. Each patient’s cancer differs significantly from all other cancers and so a safe and effective cure for each patient’s cancer will likely require a precisely-targeted custom treatment. But until recently the technologies required for crafting such custom treatments were not available, and it was widely assumed custom treatments could never be affordable.
However, since the year 2000 three new technical advances should now make it possible to provide patients with an affordable custom cancer treatment specific for their cancer.
a) One key advance is the huge reduction in the cost and great increase in the speed of sequencing the RNA transcripts present in a patient’s cancer (accessed by needle biopsy).
b) A second key advance is in computer informatics programs which now provide a fast inexpensive assessment of RNA sequence data which allows one to identify those RNA transcripts which are present in a patient’s cancer cells, but are absent from that patient’s normal cells. These “cancer-only” transcripts are primarily coded by genes which are active during embryogenesis, then deactivated before birth, but decades later reactivated due to mutation to form cancers. Precisely targeting such cancer-only transcripts is essential in order to avoid blocking transcripts present in and essential to the patient’s normal cells.
c) The third key advance is the availability of a family of drugs (Morpholinos) which can be targeted against any selected RNA transcript - designed just from knowledge of the sequence of the transcript to be targeted. For applicability against most or all cancers, such drugs: are easily and precisely targeted against any selected transcript; are completely stable in biological systems; are generally free of off-target effects; are deliverable to most or all tissues in the body; and, provide maximal versatility because they are effective in both the nucleus and the cytosol of cells.
The above technical advances open the door for development of personalized cancer treatments which: do not damage the patients; reliably destroy the cancers; and, allow precise treatment of any patient’s cancer. However, a final element is needed for development of actual drugs with these capabilities. That final element is a novel drug targeting strategy where the key elements are defined as follows and illustrated in (Figure 1).
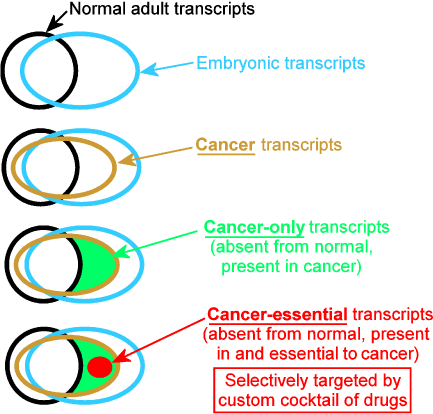
Figure 1: Functional classification of transcripts.
Transcript: An RNA sequence transcribed from a gene.
Transcriptome: All the RNA transcripts in a specified cell, tissue, or organism.
Exome: All sequences in polyA-mRNA transcripts in a specified cell, tissue, or organism.
Cancer transcript: A transcript in cancer cells.
Cancer-only transcript: A transcript present in cancers, but absent from normal adult cells (transcribed from embryo-active genes normally deactivated in adults, but reactivated in cancers, or from oncogenic viruses).
Cancer-essential transcript: A cancer-only transcript that is essential for viability of a cancer.
Custom cocktail: A combination of drugs wherein each drug is targeted against one of the cancer-essential transcripts found to be present in an individual patient’s cancer.
The way forward to custom cocktails for curing cancers in humans:
1. Develop precision-targeted drugs for most“cancer” transcripts.
a) Why RNA transcripts are the best targets.
b) Why Morpholinos are the best precision-targeted drugs.
c) May 2016: provide affordable cancer-targeted drugs to cancer researchers.
2. Use sequence information and informatics to identify “cancer-only” transcripts.
3. Use precision-targeted Morpholino drugs to identify “cancer-essential” transcripts.
4. Assemble and test custom cocktails of precision-targeted drugs for individual cancers.
a) Morpholino drugs for cell culture studies.
b) Morpholino drugs for in vivo applications.
5. Unconventional safety and efficacy trials - tailored for custom cocktails.
a) Safety trial on each Morpholino drug - with perfect antidote available.
b) Confirm “;expectation-of-efficacy” for each custom cocktail before treating patient.
6. A new era in oncology: patient’s experience from diagnosis to cure.
1. Precision-targeted drugs for “;cancer” transcripts.
a) Why RNA transcripts are the best targets.
To mount a decisive attack on multiple targets in any cancer, selection of the type of structures to be inactivated must go hand-inhand with the type of precision-targeted drugs used to achieve those inactivations.
It is also important that a great deal of information be available about the target structures, ranging from information at the submolecular level to information at the cellular, tissue, organ, and complete organism level. It is also desirable that there be detailed information about those target structures over time– preferably ranging from fertilization, through embryonic development, to birth, and on into adulthood.
I contend that RNA transcripts best meet the above criteria for target structures. We know the detailed structure of RNA at angstrom resolution, in recent years computer programs have become available to calculate the secondary structures (mainly stem/loop structures) of any known RNA sequences, and the latest sequencing methods now allow determination of the entire transcriptomes, and the much simpler exomes, of cells starting from the fertilized egg, through the complexities of embryogenesis, to birth, and into adulthood.
In contrast to RNA transcripts as the target structures, some currently-popular targets for cancer therapeutics appear to be decidedly inadequate for a decisive attack on any or all cancers. Such popular (but inadequate) targets include the following.
Replicating DNA in rapidly-dividing cells: Many chemotherapeutics indiscriminately attack replicating DNA in rapidly-dividing cells - including rapidly-dividing cancer cells. However, drugs which just attack rapidly dividing cells are notorious for devastating, and often killing, the patient in addition to the cancer. This is because the patient has a number of normal cell types which are essential to the viability of the patient and which are rapidly dividing in order to carry out their essential functions in the patient. Most notable are cells of the patient’s immune system and cells lining the patient’s intestinal track.
Receptor proteins and checkpoint proteins on cell surfaces: In recent years monoclonal antibodies have been used to target a variety of specific cell surface receptors and immune checkpoint proteins on cell surfaces that have been found to be over-expressed in some cancers. However, these classes of target structures are generally unsuitable for decisively attacking most or all cancers because such receptor proteins and checkpoint proteins are generally also present on and essential to normal cells of the patient. As a consequence, such “cancer-targeted” antibodies commonly cause substantial collateral damage to the patient. Still further, many cancers do not have such cell surface structures which are sufficiently over abundant, relative to normal cells, to make decisive targets for antibodies - leaving a large fraction of cancers un-treatable by this strategy.
Hormone receptors: Selective blocking of certain hormone receptors present on and essential to some cancers often give excellent results in the short term, but can be of limited or no value in the longer term because the high mutation rates common to cancers can result in new mutated offshoots of the original cancer which are no longer suppressed by hormonal deprivation therapy. Further, only a modest subset of cancers contains suitable hormone receptors which are essential to the growth of the cancer - leaving a large fraction of cancers un-treatable by this strategy.
Thus relative to currently-popular therapeutic targets in cancers, I believe RNA transcripts constitute far better targets for precisiontargeted drugs designed for treating any or all cancers.
b) Why Morpholinos are the best precision-targeted drugs.
The drugs to be used for a decisive attack on any cancer should be easily designed, rapidly prepared, and inexpensive to rigorously test. And those drugs must be precisely targetable against each of a multiplicity of structures essential to the viability of any patient’s cancer - all without causing significant collateral damage to that patient. And for a lasting cure, those drugs should completely destroy the entire targeted cancer before the patient’s cancer can develop resistance to the drugs.
In 1978 a new strategy for drug design was described which makes it possible to rapidly and inexpensively design and prepare precision-targeted drugs specific for virtually any selected RNA transcript, for instance: Summerton, submitted 1973, published 1979 [1]; Summerton & Bartlett, 1978a [2]; Summerton & Bartlett, 1978b [3]. This high-precision targeting exploits the Watson/Crick pairing between the four genetic letters (A pairs with T, and C pairs with G) which underlies the function of all genetic material. This antisense drug design strategy simply entails linking about 25 genetic letters into a sequence complementary to those 25 genetic letters of the RNA transcript one wishes to target. To function, such an antisense drug simply binds, via. Watson/Crick pairing, to its complementary target sequence in the selected RNA transcript - thereby inactivating its targeted RNA transcript. Alternatively, certain antisense drug types instead induce the degradation of their targeted RNA transcripts.
In principle this antisense drug design strategy is very simple.
However, in practice implementing that strategy required hundreds of scientists working for decades, funded by billions of dollars, which resulted in many dozens of inadequate structural types being synthesized and tested - before an antisense structural type was finally developed which provides the full set of key properties needed for safe and effective therapeutic use. It is noteworthy that the antisense structural type with the best combination of properties (Morpholinos) required a radical re-design of the entire backbone of natural genetic material.
But the result of that long quest was worth the decades of effort because most of the easily-designed, rapidly-prepared, precisiontargeted Morpholino antisense drugs can precisely block their specific targeted RNA transcript in a cell - generally without significantly affecting any of the more than 100,000 other gene transcripts and isoforms (splice variants) in that cell.
These non-ionic Morpholinos, now to be used for targeting cancer transcripts, have long been used for modulating the activity of tens of thousands of different gene transcripts in dozens of different species. Morpholinos were selected in part because they provide the highest sequence specificity of all gene modulating tools, reviewed in: Summerton & Weller, 1997 [4]; Summerton, 1999 [5]; Summerton, 2004 [6]; Summerton, 2007 [7].
In addition to (i) their exquisite sequence specificity, Morpholinos also: (ii) provide high efficacy (because of their high affinity for complementary RNA); (iii) are completely stable in biological systems (because of their non-ionic backbone structure); (iv) are generally free of the multiple off-target effects which commonly plague the ionic siRNA and S-DNA antisense structural types (because Morpholinos lack anionic charges on their backbones they do not interact with the many cellular factors which interact with poly-anionic nucleic acids such as siRNAs); (v) have excellent aqueous solubility (because of their high propensity for base-stacking - in contrast to the poor aqueous solubility of other non-ionic antisense structural types such as PNAs); and, (vi) provide the most predictable targeting of all antisense structural types (because of their straight-forward stericblock mechanism of action and their ability to efficiently invade the ubiquitous stem/loop structures characteristic of nearly all singlestranded RNAs).
Morpholinos also readily pass through nuclear pores. As a consequence, they are quite versatile because: (vii) they are very effective for precisely blocking translation of targeted messenger RNAs in the cytosol; and, (viii) they can precisely alter splicing of pre-messenger RNAs in the cell nucleus - enabling the reactivation of certain cancer suppressor transcripts.
It should be appreciated that precision alteration of splicing has been found invaluable for treatment of muscular dystrophy, spinal muscular atrophy, thalassemia, progeria, and many other conditions which can be corrected by altering splicing. In this context, it is noteworthy that late in 2015 a splice-correcting Morpholino for treating muscular dystrophy successfully completed a Phase 3 clinical trial and is pending regulatory approval by the FDA.
Morpholinos are also commonly used for: (ix) blocking microRNAs; (x) blocking the sequences in mRNAs targeted by microRNAs; and, (xi) blocking other non-coding RNAs.
The most challenging of all antisense applications is in the study of developing embryos where intricate cascades of gene activations and deactivations are precisely controlled with respect to both time and position in the rapidly maturing embryo. For studies in such a tremendously complicated system it is essential that the antisense oligos: (a) provide exquisite specificity for their targeted RNA; (b) achieve thorough inactivation of their targeted RNA; (c) are largely free of off-target effects; and, (d) remain stable in biological systems throughout multiple days of embryonic development. Because Morpholinos provide this unmatched combination of compelling advantages they are the only gene-modulating tools which can routinely provide reliable results in developing embryos (particularly frogs and zebrafish). For this reason, since the year 2000 Morpholinos have become the essential tools for most researchers in developmental biology, and the use of Morpholinos has revolutionized that very challenging field of research [8-10]. Scientists using Morpholinos have published over 7,500 research papers (searchable at: pubs.genetools. com) wherein these precision tools played a key role in the reported experiments, and over half of those publications came from developmental biology researchers.
c) May 2016: affordable precision cancer-targeted drugs available to researchers.
In the past decade researchers have also begun to use Morpholinos for studies of cancers, and have published well over a hundred papers reporting their results (searchable at: pubs.gene-tools.com). However, in contrast to developmental biology, where only Morpholinos are adequate for the very demanding task of gene modulation in developing embryos, in the field of cancer research most researchers instead choose to get by with the somewhat cheaper siRNAs. Thus, in the field of cancer research use of siRNAs is prevalent - in spite of siRNAs being plagued by: (i) Substantially poorer sequence specificity (because their 11-base guide sequence recognizes too little sequence information in the targeted RNA transcripts); (ii) limited stability in biological systems (because siRNAs are degraded by nucleases both outside and inside of cells); (iii) substantially poorer targeting predictability (because high complementarity to the targeted RNA transcript leads to cleavage of the targeted RNA, while lesser complementarity can lead to substantial, but unpredictable, blockage of a multiplicity of partially-complementary non-targeted RNA transcripts); (iv) inability to modify splicing (because the siRNA/ RISC complexes are located in the cytosol); and, (v) off-target effects in vivo, including interferon induction and anaphylactic shock (due to inadvertent activation of the innate immune system).
To encourage cancer researchers to use the substantially more specific, more versatile, far more stable, and much safer Morpholinos, in May 2016 GENE TOOLS, LLC will provide a catalog of precisiontargeted Morpholinos specific for many of the cancer transcripts and most of the cancer-only transcripts in humans. These will be available at a specially reduced price of $250 for an amount sufficient to treat cells in 500 wells of 96-well cell culture plates (0.2 ml/well). This will put the cost of such Morpholinos below the cost for a corresponding amount of the less specific, less stable, and less versatile siRNAs. Thus, researchers in oncology will be able to enjoy all the compelling advantages of Morpholinos long enjoyed by researchers in developmental biology - but at a price even lower than for siRNAs.
2. Use sequence information to identify “cancer-only” transcripts.
A key challenge for cancer therapeutics has been to achieve complete destruction of the cancer without also damaging or killing the patient. The obvious solution to this challenge is to target a structure: a) which is essential to the viability of the patient’s cancer cells; but, b) is absent from the patient’s normal cells.
Before completion of the Human Genome Project in 2003 identifying structures present in a given patient’s cancer, but absent from that patient’s normal cells, was a formidable task. However, over the last 15 years great increases in speed and great reductions in costs of genetic sequencing have been achieved - such that now RNA transcripts present in a patient’s cancer cells and absent from that patient’s normal cells can be routinely identified in hours to a few days for less than a thousand dollars.
Since this blossoming of genetic sequencing and ultra-rapid analysis of the resulting sequence information, scientists have surprisingly learned that most or all cancers have a large number of RNA transcripts which are normally present only during embryogenesis, then are normally deactivated before or shortly after birth, but then decades later (due to random mutations, infection with oncogenic viruses, or other insults) genes coding for those “embryonic” transcripts are abnormally reactivated - leading over the course of years to the multiple abnormal and uncontrolled properties of cancers [11-13].
The probable explanation for this was well described in 2007 by Robert Weinberg [14] in an interview for the MIT Technology Review. In essence, Weinberg made the case that cancers (over the course of many years and through multiple random mutations) cobble together the complex set of behaviors characteristic of cancers - simply by randomly resurrecting key embryonic behavioral programs. In that way, with a few random activation and deactivation steps in key genes in emerging cancer cells, under selection pressures (and probably with a great many failures) one or more cancer cells can stumble, over time and in a number of stages, into a capability to modulate the expression of whole cohorts of responder genes (especially transcription factors) that allow the cancer to effectively go wild - until the cancer’s lack of control destroys the patient.
In recent years this connection between embryonic behavioral programs and cancers has gained a great deal of experimental support. Thus, it seems very likely that by sequencing the transcriptome (or just exome) of a patient’s cancer, that cancer will be found to contain some combination of multiple RNA transcripts that are: a) present in the cancer; but, b) absent from normal cells of the patient. While the exact set of such “cancer-only” transcripts will likely vary wildly from cancer to cancer, I believe that together all the cancer-only transcripts in humans constitute by far the best pool of drug targets for a family of safe, effective, and affordable precision-targeted Morpholino drugs for curing most or all cancers.
3. Use Morpholino drugs to identify “cancer-essential” transcripts.
While targeting just cancer-only transcripts will prevent damage to the patient, nonetheless, I believe many of those cancer-only transcripts identified by transcriptome sequencing and informatics analysis will not be good therapeutic targets because they are not essential to the viability of the cancer (i.e., many are passengers instead of drivers). Thus, after identifying most or all of the canceronly transcripts in humans, it will likely be necessary to next identify those particular cancer-only transcripts which are also essential to the viability of the cancers, (either singly or possibly in combination with other cancer-only transcripts). Cancer-only transcripts in this special “essential” subset are designated: “cancer-essential” transcripts.
I believe identification of cancer-essential transcripts in a cancer can best be achieved by testing, in cells cultured from that cancer, precision-targeted drugs specific for each of the cancer-only transcripts found (via. sequencing) to be present in that cancer. In May 2016 GENE TOOLS intends to make available to the oncology research community an arsenal of precision-targeted Morpholinos effective against most or all of the known cancer-only transcripts in humans (estimated to be a few hundred to a thousand cancer-only transcripts).
Experimental identification of cancer-essential transcripts for that cancer will likely entail:
1. Take cells from a cancer and have the cancer’s polyA-mRNA transcriptome sequenced.
2. From the resultant sequence information use a suitable informatics program to identify the cancer-only transcripts in that cancer.
3. Also from the initial biopsy, or other sampling of the cancer cells, culture the cancer cells and plate them out in 96-well culture plates.
4. Expose each well of the cultured cancer cells to one of the precision-targeted drugs specific for one of the cancer-only transcripts identified by sequencing that cancer. Later it may be necessary to test these drugs in combinations in order to achieve definitive killing of the cancer cells.
5. A well where all the cancer cells are killed indicates the precision-targeted drug or drug combination in that well was targeted against a cancer-essential transcript or canceressential set of transcripts. A well where the cancer cells remain viable indicates the precision-targeted drug or drug combination in that well was not targeted against a canceressential transcript or set of transcripts (ie., the target was not a driver, but instead only a passenger target).
While a number of controls will also be needed, as well as further studies to confirm the initial results, nonetheless, this simple test strategy should indicate which of the cancer-only transcripts constitute the key cancer-essential transcripts in that particular cancer.
As noted earlier, an exome comprises all the sequences of mature messenger RNAs in a specified cell, tissue, or organism. But the exome comprises just a small subset of the corresponding transcriptome, which also includes a host of non-coding transcripts. Initially our focus will be only on the exome of normal adult cells and the exome of selected cancers, or more specifically, the polyA-mRNA transcriptomes of normal adult cells and of selected cancers. Focusing just on these polyA-mRNA transcriptomes will substantially simplify the sequencing process and greatly reduce the complexities of the subsequent informatics assessment of the sequence results. My expectation is that exome information alone will be adequate for developing safe and effective custom cocktails for most or all cancers. But in rare cases it may be necessary to utilize sequence information from full transcriptomes.
4. Assemble and test custom cocktails for individual cancers.
Once a substantial number of cancer-essential transcripts have been identified, and the cancer-essential transcripts present in a given patient’s cancer have been determined by sequencing, the pieces will be in place to assemble a custom cocktail. Probably about 2 to 6 of the precision-targeted drugs specific for cancer-essential transcripts present in that patient’s cancer will provide a custom cocktail effective to completely destroy that cancer - with no harm to the patient, see also: Summerton [15].
A second round of cell culture experiments can then quickly and inexpensively assess a variety of prospective cocktails to determine how many drugs and which drugs are optimal for achieving complete destruction of that particular cancer. With increasing experience, design of such custom cocktails should become fast and reliable.
a) Morpholino drugs for cell culture studies.
Most of the early studies with precision-targeted Morpholinos will be focused on determining which of the many cancer-only transcripts constitute the special subset comprising the canceressential transcripts. These studies will be carried out with cultured cancer cells where the culture medium typically contains 10% serum. For such studies with cultured cells the Morpholino drugs will include a new 4-component delivery system that is both very effective and quite safe for use with cultured cells in culture medium containing up to 10% serum. In May of 2016 GENE TOOLS plans to introduce this new cultured-cell delivery system for cancer-targeted Morpholino oligos - which should serve for identification of many or most of the cancer-essential genes in humans. That information alone should be of tremendous value to the entire oncology research field.
b) Morpholino drugs for in vivo applications.
Once the subset of cancer-essential transcripts have been identified, it will be a simple matter to assemble a variety of custom cocktails for any given cancer, and then study in cell cultures just which custom cocktails are most effective. Once that has been done with cultured cancer cells, the next stage will entail use of those cocktails in living animals. Initially this will likely entail testing the cocktails against human cancers in SCID (severe combined immunodeficiency) mice.
Success in that mouse system will be followed by safety trials in healthy humans wherein each precision-targeted Morpholino drug specific for one of the cancer-essential transcripts is tested for safety. Thereafter, a small number of cancer patients will enter single-person efficacy trials wherein each such trial will entail injection of one custom cocktail into the cancer patient for which that cocktail was designed - where the composition of that cocktail is selected from precision-targeted Morpholinos, which have proven safe in a safety trial, and where the component Morpholino drugs are selected based on the sequence information from that patient’s cancer.
For those in vivo studies the Morpholinos will be formulated with a different fourth delivery component than used in the cell culture studies. This is because the fourth component in the delivery system used for cultured cells does not work well in the presence of the high serum concentration present in the blood of mammals. We are currently working to modify that 4th component to make it compatible with the high serum concentration it will face in vivo. GENE TOOLS hopes to introduce such a 4-component in vivo delivery system before the end of 2016.
5. Unconventional safety and efficacy trials - tailored for custom cocktails.
The FDA’s rules and regulations were developed with the objective of assuring that drugs developed and used in our nation will likely provide benefits that outweigh the likely risks. This fundamental objective of drug developers and regulators is both reasonable and highly desirable and should be met for all drugs to be used in humans.
But in an effort to achieve this highly desirable objective, very complex rules and regulations have been adopted by the FDA which result in very long times (one to two decades) and exorbitant costs (hundreds of millions to billions of dollars) for developing and testing a breakthrough drug. It appears those long times and exorbitant costs are the collateral damage that goes with the FDA’s need to regulate development of a huge array of drugs ranging from treatments for mild conditions (such as insomnia) to treatments for ravaging lifedestroying diseases (such as cancers and lethal viruses).
However, in the case of personalized drugs, or custom cocktails of drugs, the FDA’s current very-long-time/very-high-cost route to regulatory approvals appears to effectively preclude development of most or all personalized drugs and custom cocktails of drugs.
Therefore, for society to reap the likely huge benefits of personalized drugs and custom cocktails of drugs for treating severe and life-threatening diseases, it will be necessary to provide solid evidence that the drugs’ will likely provide benefits that outweigh their likely risks - and to demonstrate this fast enough that the patient to receive such a personalized drug or custom cocktail has not died waiting for the treatment. And demonstration of safety and efficacy should not cost so much for regulatory approval that it bankrupts our healthcare system.
The following proposals for safety and efficacy trials for the custom cancer treatments described herein are tailored to assess safety and provide a compelling expectation-of-efficacy sufficient for regulators to make an informed decision on whether or not these custom treatments will likely provide benefits that outweigh the likely risks.
a) Safety trial on each Morpholino drug - with perfect antidote available.
While adverse effects are far less common with Morpholino drugs than with any of the other antisense types (one of the principal reasons why Morpholinos are the only antisense structural type commonly used in developmental biology studies), nonetheless, adverse effects from certain Morpholino drugs can occur on rare occasions. Therefore, it is desirable that each of the Morpholino drugs targeted against cancer-essential transcripts (the component drugs in custom cocktails) be first tested individually for safety in humans.
Happily for the special case of antisense drugs, the risk in any such safety trial can be dramatically reduced by providing a means to quickly, thoroughly, precisely, and affordably halt the activity of that drug within the patient. Such a means comprises the sense oligo exactly complementary to the antisense drug. Specifically, in the very rare case where a Morpholino antisense drug causes an adverse effect in a treated subject, the exact complementary sense oligo (the perfect antidote [16]) can be quickly delivered into the subject. That sense oligo, on encountering the antisense drug, will quickly pair with the antisense drug to form an inert duplex, thereby inactivating the antisense drug and halting the undesired adverse event. This riskreduction strategy is illustrated in (Figure 2).
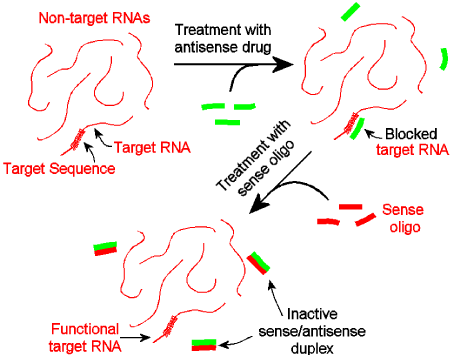
Figure 2: Risk- reduction method.
Designing such perfect antidotes takes only a few seconds, and the risk-reducing sense oligo can be synthesized at the same time as its complementary antisense drug. While such a sense oligo would only be used in the very unlikely event an antisense drug caused a significant adverse event in a test subject, it should be appreciated that simply having such a sense oligo immediately available during the safety trial provides a tremendous risk-reduction benefit to the test subject by allowing a rapid and specific inactivation of the antisense drug within the patient should such a need ever arise.
b) Confirm “expectation-of-efficacy” for each custom cocktail before treating patient.
When a patient is found to have a cancer, the clinician will take a needle biopsy or otherwise collect a sample of the cancer cells. Part of that cancer sample will be sent out for sequencing of the exome (or transcriptome), and part will be grown in culture medium and then those cancer cells added to tissue culture plates. Once a listing of the cancer-essential transcripts in that patient’s cancer arrives from the sequencing/informatics center (typically at a University or commercial lab), each well of cancer cells in the culture plate will be exposed to a Morpholino drug targeted against one of the canceressential transcripts, identified from the sequencing of the patient’s cancer. After incubation of the culture plate for a suitable period (probably about 2 days), observation of which wells contain dead cancer cells should confirm the sequencing result and so indicate
the specific set of cancer essential transcripts present in that patient’s cancer. This, in turn, will indicate which precision-targeted drugs to combine to form the custom cocktail for treating that patient. An order will then be placed for that custom cocktail. Days later when the patient’s custom cocktail arrives from the supplier an appointment will be set up for the injection.
When the custom cocktail arrives, a small portion will be used to treat the previously-cultured cancer cells to confirm that the cocktail is indeed effective to completely kill that patient’s cancer cells - thereby providing a compelling “expectation-of-efficacy” that the cocktail will be effective against that patient’s cancer. With that evidence in hand, the patient will then be injected with that custom cocktail.
6. A new era in oncology: patient’s experience from diagnosis to cure.
I envision the cancer patients experience with the custom cocktail treatment will entail two steps.
a) After a patient has been diagnosed with cancer, the clinician will collect a sample (biopsy) of the cancer cells and the patient will go home for about 10 days while those cancer cells are sequenced and cultured and an appropriate custom cocktail is ordered (based on the sequencing results).
b) After the patient’s custom cocktail arrives from the supplier and its efficacy against that patient’s cultured cancer cells is confirmed, that cocktail will be injected into the patient.
I predict patients treated with such custom cocktails will no longer suffer from the nausea, diarrhea, hair loss, high susceptibility to infections, and the host of other afflictions typically accompanying treatment with current cancer therapies. More important, these custom cocktails are expected to reliably cure most or all patient’s cancers, including late-stage metastatic cancers, thereby avoiding the devastating relapses that so commonly lead to many patients’ untimely and terrible deaths. Also of great importance to healthcare systems worldwide, I estimate that these custom cocktails will be substantially cheaper than the extensive course of treatments now used (often unsuccessfully) for treating cancers - treatments which often cost in the range of $ 50,000 to $ 100,000 per patient.
In regard to costs of such custom cocktail treatments, based on current production costs, factoring in projected cost savings from scale-up of Morpholino production, and presuming production costs are not unreasonably increased by regulatory requirements, I estimate a custom-cocktail treatment will cost between $ 15,000 and $ 30,000 per patient, depending on the number of cancer-essential genes which are targeted. There may also be an extra $ 2,000 “delivery charge” if the cancer is shielded by the blood/brain barrier and so requires a special transcytotic delivery component or other special procedure for delivery of the custom cocktail into the central nervous system. But after this treatment strategy becomes widely used these costs should drop considerably due to economies of scale and streamlining of procedures.
Regarding time to implementation, I expect a proof of principle demonstration through the sequencing and cell culture steps will be completed by mid-2016. If a reasonable number of oncology research groups soon become involved, then it should be possible to identify many or most cancer essential genes in humans by the end of 2017, and by the end of 2018 it should be possible to assemble custom cocktails for a large number of patients’ cancers. Then, baring undue delay by regulatory agencies, safe, effective, and affordable cures for any cancer, including late-stage metastatic, may be broadly available to patients by or before 2020.
References
- Summerton J. Intracellular inactivation of specific nucleotide sequences: A general approach to the treatment of viral diseases and virally mediated cancers. J Theor Biol. (Submitted 1973) 1979; 78: 77-99.
- Summerton J, Bartlett PA. Sequence-specific crosslinking agents for nucleic acids. Use of 6-bromo-5,5-dimethoxyhexanohydrazide for crosslinking cytidine to guanosine and crosslinking RNA to complementary sequences of DNA. J Mol Biol. 1978; 122: 145-162.
- Summerton J, Bartlett P. Nucleic acid crosslinking agent and affinity inactivation of nucleic acids therewith. 1978; US Patent 4,123,610
- Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997; 7: 187-195.
- Summerton J. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim Biophys Acta. 1999; 1489: 141-158.
- Summerton J. Morpholinos and PNAs compared. Lett Pep Sci. 2004; 10: 215- 236.
- Summerton JE. Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Curr Top Med Chem. 2007; 7: 651-660.
- Ekker SC. Morphants: a new systematic vertebrate functional genomics approach. Yeast. 2000; 17: 302-306.
- Heasman J. Morpholino oligos: making sense of antisense? Dev Biol. 2002; 243: 209-214.
- Special Issue: Morpholino Gene Knockdowns - all 27 research reports in the July issue of the journal: Genesis. 2001; 30: 89-200.
- Kho A, Zhao Q, Cai Z, Butte A, Kim J, Pomeroy S, et al. Conserved mechanisms across development and tumorgenesis revealed by a mouse development perspective of human cancers. Genes and Development. 2004; 18: 629-640.
- Naxerova K, Bult C, Peaston A, Fancher K, Knowles B, Kasif S, et al. Analysis of gene expression in a developmental context emphasizes distinct biological leitmotifs in human cancers. Genome Biology. 2008; 9: 108.
- Barrett C, De Boever C, Jepsen K, Saenz C, Carson D, Frazer K. Systematic transcriptome analysis reveals tumor-specific isoforms for ovarian cancer diagnosis and therapy. Proc Natl Acad Sci USA. 2015; 112: 3050-3057.
- Bourzac K. How a tumor is like an embryo. MIT Technology Review. 2007.
- Summerton JE. Custom cancer therapies: safe and effective treatments for most or all cancers. Ann N Y Acad Sci. 2003; 1002: 189-196.
- Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, et al. RNA aptamers as reversible antagonists of coagulation factor IX a. Nature. 2002; 419: 90-94.