
Research Article
Austin Food Sci. 2022; 8(1): 1053.
Analysis of Influencing Factors on Milk Production Traits of Chinese Holstein A2 Cattle and Proposal of a Threedivision Rearing Strategy
Haitong Wang1*; Chunfang Li1,2; Liangkang Nan1; Yikai Fan1; Xiangnan Bao3; Xinxin Zhang1; Jingjing Zhang1; Chu Chu1; Yun Liu1; Bo Song2; Shengchao Liang2; Chendong Yang2; Guochang Yang1; Yabin Ma2; Junqing Ni2; Wei Sun3; Xihe Li3; Shujun Zhang1
¹Key Lab of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education, Huazhong Agricultural University, China
²Hebei Livestock Breeding Station, China
³National Center of Technology Innovation for Dairy Industry, China
*Corresponding author: Haitong Wang, Key Lab of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education, Huazhong Agricultural University, Wuhan 430070, China. Email: htw0411@webmail.hzau.edu.cn
Received: October 04, 2024; Accepted: October 25, 2024; Published: November 01, 2024
Abstract
Casein (CN) is an important factor determining the nutritional level of milk. A2 milk has been highly popular worldwide for its higher nutritional value and easier digestion and absorption than non-A2 milk. Given the burgeoning demand and economic advantages associated with A2 milk, breeding specialists have embarked on selecting A2 cows from dairy herds by genotyping the Beta Casein (β-CN) encoding genes in dairy cows to form a specialized A2 dairy herds and produce A2 milk. In this study, the milk production traits of Chinese Holstein A2 cows were analyzed in relation to lactation days and parity. Based on these analyses, a three- division rearing strategy was proposed, in which cows should be separated and reared according to genotype, parity and lactation stage. Initially, A2 and non-A2 cows were grouped and A2 cows were found to produce 5 types of milk: protein-rich A2 milk starting from day 91 of the second parity, high-β-CN A2 milk from day 120 of each parity, low-lactose A2 milk during the first 30 days of the second and subsequent parities, low-fat A2 milk between days 31 to 90 of each parity, and regular A2 milk during the remaining lactation periods. Non-A2 cows exhibited a higher milk yield, capable of producing a greater quantity of milk that meets national fresh milk standards. Economically, segregating A2 from non-A2 cows into distinct herds proved more beneficial than mixed-herd rearing. By optimizing cow utilization, our strategy will ensure both milk yield and milk quality, meet the diversified needs of consumer groups for dairy products, and increase the economic benefits of dairy farms.
Keywords: A2 milk; Beta-casein; Chinese Holstein cows; three-division rearing strategy
List of Abbreviations
a-LA: Alpha-Lactalbumin; as1-CN: Alpha (s1)-Casein; as2-CN: a- (s2)- Casein; β-CN: beta-casein; β-CM7: β-casomorphin-7; β-LG: β-Lactoglobulin; CN: Casein; DHI: Dairy Herd Improvement; DIM: Days in Milk; FY: Fat Yield; HPLC: High-Performance Liquid Chromatography; IEF: Isoelectric Focusing; κ-CN: kappa-Casein; MIR: Mid-Infrared; MY: Milk Yield; PY: Protein Yield; RMSE: Root Mean Square Error; SCC: Somatic Cell Counts; SCS: Somatic Cell Score; SNF: Solid Non Fat; TMR: Total Mixed Ration; TS: Total Solids
Introduction
The proteins in milk are the determinants for the nutritional properties of milk, and milk casein and whey protein as well as their ratio play an important role in the production of liquid milk, cheese, and other dairy products. Casein (CN) accounts for about 70-80% of the total milk protein, an is further categorized into Alpha (s1)- Casein (as1-CN), Beta-Casein (β-CN), Alpha (s2)-Casein (as2-CN), and Kappa-Casein (κ-CN) [1]. In recent decades, the effect of Single Nucleotide Polymorphisms (SNPs) of milk protein-encoding genes on milk protein content and functions have been well studied [2- 4]. Beta-casein, accounting for 30-40% of the total casein content, is encoded by the CSN2 gene with up to 15 genetic variants, of which A1 and A2 are the two most common variants with variant A2 being a wild type [5,6]. The only difference between A2 and A1 lies in amino acid 67. The generation of A1 is due to a mutation (C → A, CSN2 X14711 8101) at position 8101 in the CSN2 gene (GenBank, accession number: NC_037333). This mutation results in the conversion from A2 gene's codon CCT to CAT and the replacement of Pro with His in the coding product, eventually forming the A1 variant [7]. A1-type and A2-type of β-CN can be separated and identified by multiple methods such as Isoelectric Focusing (IEF) and High-Performance Liquid Chromatography (HPLC) [8,9].A1-type and A2-type β- CN can be decomposed by digestive enzymes to produce different bioactive peptides, and the β-casomorphin-7 (β-CM7) produced by type A1 reduces gastrointestinal motility, decreases gastrointestinal immune regulation, and promotes inflammation [10]. A2 milk and the corresponding dairy products containing A2-type β-CN can improve milk intolerance reactions, alleviate symptoms such as flatulence in patients [10]. Thus, A2 milk is highly popular among consumers, and its consumption increases rapidly with a higher price than other milk. In traditional small-scale dairy farming, the entire herd is often treated as a single rearing group, offering the advantages of simplicity, efficiency, and labor-saving. This approach allows for uniform rearing without distinguishing between the diets and management of lactating cows and those at other stages, thereby significantly conserving labor. However, the nutritional needs of cows vary with different stages, particularly during lactation, a critical period for milk production which is a primary economic contributor to dairy farms. Appropriate nutrition can notably enhance milk yield and quality. Consequently, for large-scale, intensive dairy farms, segmenting herd management is a fundamental and crucial task. The effectiveness of this segmentation can directly influence the profitability of the dairy farm. In practical operations, dairy cattle are typically categorized into groups such as calves, heifers, and mature cows (including peripartum, freshly calved, dry, and lactating cows).
To meet consumer’s demand for A2 milk and improve its production efficiency, molecular biology techniques such as gene sequencing have been used to detect and screen A2 cows carrying base CC (Pro, also known as A2A2) to form A2 dairy herds for exclusive production of A2 milk containing non-allergenic A2 type β-CN. A1 cows carrying base AA (His, also known as A1A1) and A1A2 cows carrying base AC are gathered together to form a non-A2 cow herd to produce conventional milk (designated as non-A2 milk). Typically, the proportion of A2 and non-A2 cows in a mixed herd is about 30% and 70%, respectively [11], and some farms rear A2 cows and non-A2 cows in separate herds [12,13].
The milk production traits of dairy cows are influenced by a variety of factors, including genetic, health, and nutritional and nonnutritional elements [14-18]. Studies highlighted significant effects of parity and lactation on these traits [1-,20]. However, in Chinese Holstein A2 cows, the understanding of how milk yield, composition, and protein fractions vary with lactation stages and parities, along with optimal strategies for A2 milk production, remains limited. Therefore, the objectives of this study within the Chinese Holstein dairy cow population are to: (1) analyze the patterns of variation in milk yield, milk composition, and protein fractions of A2 cows across lactation and parity; (2) investigate effective production strategies for A2 milk.
Materials and Methods
Farm data and sample collection
The Animal Management and Ethics Committee of Huazhong Agricultural University reviewed and approved the experimental protocol for this project (HZAUCA-2020-0001). This study was conducted with a lactating herd of Chinese Holstein dairy cows housed in 2 dairy farms (Farm A and Farm B) in northern China. Cows were raised in house (about 18 hour per day) and in yards (about 6 hour per day) and fed on Total Mixed Ration (TMR) with free access to water. Milk samples were collected once a month from all animals during the morning milking (4:00-6:00 am) from November 2020 to April 2023 with 45 ml per each sample. After sample collection, 0.35 μL of Bropol preservative was added to the sampling bottle and thoroughly mixed with milk prior to the delivery to DHI (dairy herd improvement) Laboratory. The routine milk composition determination was performed within 24 hours after sample collection. A total of 38,641 milk samples were collected from 4,781 healthy lactating cows (excluding sick cows with clinical signs or under treatment), and 38,641 DHI data were collected.
Standardization of instruments
The data obtained from the milk composition analyzer were standardized according to the monthly standards of milk fat and milk protein percentages, the quarterly standards of lactose percentage prepared by the National Animal Husbandry Administration of China, and quarterly standards of urea nitrogen developed by the ChemSpec 150 (Bentley, Minnesota, USA).
Determination of milk components
Milk components including fat (%), protein (%), lactose (%), SNF (solid non fat, %), TS (total solids, %) were measured with Milkoscan FT+ (Foss, Hilleroed, Denmark) and Bentley FTS (Bentley, Minnesota, USA).Mid-infrared (MIR) spectroscopy data were used for protein fraction prediction. Somatic cell counts (SCC) were obtained from Fossomatic FC counter (Foss, Hilleroed, Denmark) and SOMACOUNT FCM (Bentley, Minnesota, USA) and converted to somatic cell score (SCS) according to the formula: SCS = [log2(SCC/100) + 3] [21].
Prediction of protein fraction contents
The reference values of contents of 5 protein fractions (β-CN as1-CN κ-CN a-LA and β-LG) and total casein were determined with a Waters high performance liquid chromatograph (2695) by previously reported RP-HPLC method [8]. To predict the content of each of above-mentioned 6 proteins, corresponding prediction models were developed based on the mid-infrared data. The R2 of cross-validation sets of these 6 models was 0.7418, 0.7939, 0.8382, 0.7794, 0.7687, and 0.7501 with the corresponding Root Mean Square Error (RMSE) of 2.1187, 1.3505, 0.4754, 0.35470.5249 and 5.0167 g/L, respectively (unpublished).
Screening of valid samples and data
A total of 38,641 DHI data with the information on cow's parity,days in milk (DIM), and milk yield were collected. Fifteen traits of lactating cows were investigated, including 7 traits obtained from DHI reports, namely, daily milk yield (MY, kg/d), protein content (%), fat content (%), lactose content (%),SNF (%), TS(%) and SCS, 2 traits derived from content multiplied by milk yield(fat yield and protein yield, kg/d) and 6 traits predicted by models, namely, total casein (caseins, g/L), β-CN (g/L), aS1-CN (g/L), κ-CN (g/L), a-LA (g/L),and β-LG (g/L) content. The 2,937 measured cows from farm A yielded 18,870 DHI data, and a total of 19,971 DHI data were obtained from farm B with 1,844 cows.
The abnormal values beyond the detection range were excluded from the raw data, and samples with milk fat percentage of 1.5~9%, milk protein percentage of 1~7%, and SCC of 0~1 million/ml were retained. Ultimately, 29,736 valid data were obtained from 2,956 cows, including 12,588 records of 1,470 cows from farm A and 17,148 records of 1,486 cows from farm B. The parity range was 1 to 7 and DIM range was 0 to 400 d. There were 5 levels of parity (1, 2, 3, 4, =5) and 12 levels of DIM with 30d intervals (0~30d, 31~60d, 61~90d, 91~120d, 121~150d, 151~180d, 181~210d, 211~240d, 241~270d, 271~ 300d, 301~330d, >330d).
Statistical Analysis
A mixed linear model was used to assess the contribution of the parity and DIM to the phenotypic variance of traits:
yijklm=μ+Parityi+DIMj+SCSk+Parity*DIMl+ Herd-datem+ eijklm [1]
Where yijklm the response of the trait (milk yield, fat yield, protein yield, protein content, fat content, lactose content, SNF, TS, total casein, β-CN, aS1-CN, κ-CN, a-LA and β-LG contents); μ is the overall mean for each trait; Parityi is the fixed effect of parity at the ith level (i=1, first parity; i=2, second parity; i=3, third parity; i=4, fourth parity; i=5, fifth and subsequent parity);DIMj is the fixed effect of DIM at the jth level (j=1,2,... ,12); SCSk is the fixed effect of SCS at kth level (k= 1, SCS = -1; k=2, -1 < SCS = 0; k=3, 0 < SCS = 1; k=4, 1 < SCS = 2; k=5, 2 < SCS = 3; k=6, 3 < SCS = 4; k=7, 4 < SCS = 5; k=8, SCS > 5); Parity*DIMl is the interaction effect of parity and DIM; Herd-datem is the random effect of the mth herd-test day (m=1-22); eijklm is the random residual, (assumed to follow eijklm~N (0, se 2) normal distribution where se 2 is the residual variance).
Results and Discussion
Milk Yield, Milk Composition and Protein Fraction of A2 Cows
The descriptive statistics of investigated traits were presented in Table 1. The average daily milk yield (36.56 kg/d), daily protein yield (1.24 kg/d) and protein content (3.41%) of cows in this study was similar to that reported before [3,12,22]. And the content of fat and lactose was 3.87% and 5.14%. Among the casein fractions, the highest content was as1-CN (10.60 g/L), followed by β-CN (8.79 g/L), and the lowest content was κ-CN (4.16 g/L); the content of β-LG (3.14 g/L) was higher than that of a-LA (0.96 g/L) in whey protein.
Item1
Records
Mean
SD
CV
P52
P952
Yield, kg/d
Milk yield
29,736
36.56
12.90
0.35
15.0
57.8
Protein yield
29,736
1.24
0.44
0.35
0.52
1.97
Fat yield
29,736
1.40
0.58
0.41
0.55
2.42
Milk composition, %
Protein
29,736
3.41
0.39
0.11
2.83
4.08
Fat
29,736
3.87
0.88
0.23
2.51
5.38
Lactose
29,736
5.14
0.29
0.06
4.68
5.51
TS
29,736
13.50
1.90
0.14
11.22
17.65
SNF
29,736
9.47
1.14
0.12
8.38
12.35
SCS, units
29,736
2.77
1.60
0.58
0.26
5.12
Protein fraction, g/L
T-CN
15,359
25.02
9.56
0.38
7.40
37.86
β-CN
14,276
8.79
4.63
0.53
2.90
15.42
as1-CN
15,692
10.60
4.75
0.45
5.68
20.93
κ-CN
15,698
4.16
1.42
0.34
2.17
6.21
a-LA
15,680
0.96
0.25
0.26
0.57
1.36
β-LG
15,488
3.14
1.52
0.48
0.91
5.77
1SCS: somatic cell score, SCS = log2(SCC/100) + 3
2P5: 5th percentile; P95: 95th percentile
Table 1: Descriptive statistics for milk yield, milk composition and protein fraction of A2 cows.
Effects of DIM and Parity on Milk Production Traits of A2 Cows
Parity and DIM are important factors affecting the phenotypes of multiple traits [3,22]. In the present study, DIM has a significant effect on all traits, and parity has significant effects on milk yield, protein yield and fat yields and contents of protein, lactose, κ-CN and β-LG.
The yield of milk, protein and fat varied in a zenith curve with lactation days (Figure 1A): daily milk yield gradually increased, reached a peak on day 31-60, and maintained until about day 90, followed by a slow decrease until the end of lactation, with higher yields on days 0-120, which was in line with the previous report [23]. The effects of the parity of the cows in this study also confirm the expected increase in yield in the 1st to 3rd lactation and gradually decreased in the 3rd to 5th lactation and thereafter (Figure 1B). Protein yield was significantly higher in the 2-4th lactation than in the other 2 lactations (P<0.05), which was in accordance with that has been previously reported [24]. The curves of protein content and fat content were nadir curve, which were inversely related to yield. The protein and fat content fell from about day 31-60, and then rose in day 151-180, and they were highest in the second parity. Lactose content varied slightly over the entire lactation, slowly ascending, reaching the maximum at day 31-120, then slowly descending, and it slowly decreased with increasing parity. The change trends of SNF and TS with DIM were similar to those of protein content, since protein was one of the important solid component in milk. The variations in SNF and TS with parity are not significant, with a higher content observed in the parity 2-3 (Figures 2A and 2B).
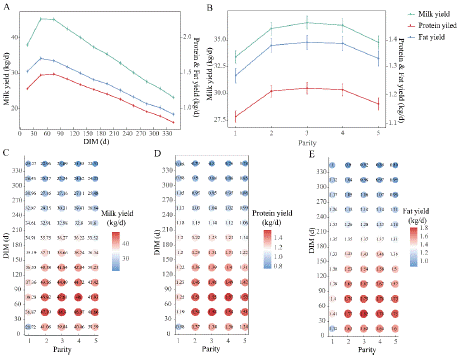
Figure 1: Effects of DIM and Parity on daily yield of milk, protein and fat. (A)
Effect of DIM on milk yield, protein yield and milk fat yield; the primary y-axis
is the range of milk yield, and the secondary y-axis is the range of protein
yield and milk fat yield; (B) Effect of parity on daily yield; (C) Combined effect
of DIM and parity on milk yield; (D) Combined effect of DIM and parity on
protein yield; (E) Combined effect of DIM and parity on fat yield.
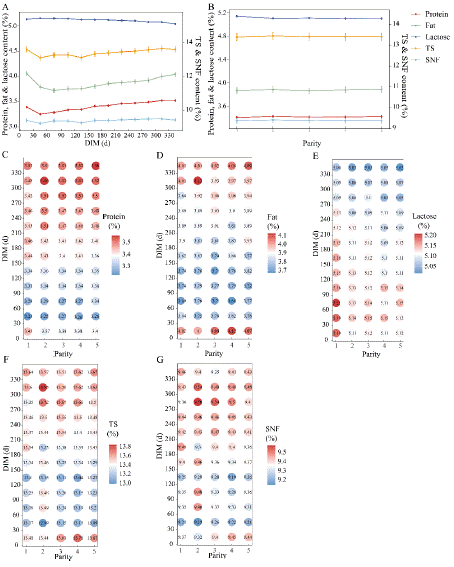
Figure 2: Effects of DIM and parity on milk composition.(A) Effect of DIM
on protein, fat, lactoes, TS and SNF content; the primary y-axis is the range
of protein, fat and lactose content, and the secondary y-axis is the range of
TS and SNF content; (B) Effect of parity on milk composition; (C) Combined
effect of DIM and parity on protein content; (D) Combined effect on fat
content; (E) Combined effect on lactose content; (F) Combined effect on TS
content; (G) Combined effect on SNF content.
In terms of protein fractions, total casein content and β-CN content followed the same trend as protein content throughout lactation, showing a curve of first decreasing and then increasing. Total casein content was higher during the 1st and 2nd parities, while β-CN content peaked during the 3rd and 4th parities. The production of milk protein was highest during the 2nd to 4th parities, resulting in the peak total casein during the 2nd parity and the highest β-CN production during the 3rd and 4th parities. The κ-CN level increased gradually throughout lactation and reached the maximum at the second parity. as1-CN, a-LA and β-LG showed smoother curves than other traits (Figure 3A and 3B).
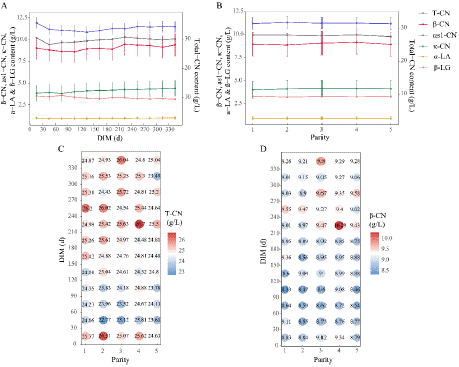
Figure 3: Effects of DIM and parity on protein fractions. (A) Effect of DIM
on β-CN, as1-CN, κ-CN,a-LA, β-LG and total-CN content; the primary
y-axis is the range of β-CN, as1-CN, κ-CN, a-LA and β-LG content, and
the secondary y-axis is the range of total-CN content; (B) Effect of parity on
protein fractions; (C) Combined effect of DIM and parity on total-CN content;
(D) Combined effect on β-CN content.
Effective production strategies for A2 milk
Compared to non-A2 milk, A2 milk exhibited higher levels of protein content, TS, and SNF, alongside lower concentrations of fat and lactose, as well as hypoallergenic β-casein. In practical dairy production, DIM and parity are typically interactive factors influencing milk yield traits. Therefore, a production strategy for A2 milk was proposed, based on the interactive effects of DIM and parity on milk production.
Milk yield, protein yield, and fat yield were higher within approximately day 0-150 of lactation and peaked within day 31-90 of lactation at the second to fourth parity (Figure 1C~1E). The protein content decreased from day 31 to day 60, subsequently increased, and then returns to the levels observed during the initial 0-30 days in the period of 61-90 days. Furthermore, protein content reached the highest values at the second parity. SNF and TS were higher in the late lactation than in the early and middle lactation and peaked at the second parity (Figure 2F and 2G). β-casein content was relatively low during the initial 0-30 days and the first lactation. In the mid to late stages of lactation, β-CN content exceeded early lactation levels, reaching approximately 9g/L at day120. Variations across lactations are minimal, with the peak occurring around day 210-240 of the fourth lactation (Figure 3D). In A2 milk, the fat content displayed a nadir curve relative to DIM across each parity, with the nadir observed between 31-60 days, and marginal differences between lactations (Figure 2D). Lactose concentration in milk was positively correlated with milk yield, varying in tandem with production levels. Lactose is one of the main determinants of milk yield with multiple important functions. Its metabolic intermediates can serve as substrates for protein and fat synthesis, and it can also provide energy for cellular metabolism [25]. In A2 cattle, the lactose content was significantly higher during the first parity compared to subsequent ones, with the period from day 31-150 exhibiting elevated lactose levels relative to other periods (Figure 2E).
The above results implied a three-division rearing strategy was proposed that on the premise of grouping A2 cows and non-A2 cows according to their genotypes, A2 cows are further groped and reared base on DIM and parity. Protein-rich A2 milk would be produced from day 91 postpartum in the 2nd lactation, high β-CN A2 milk from day 121 in each lactation, low-fat A2 milk during days 31-90 of each lactation, low-lactose A2 milk during days 0-30 in all but the first lactation, and standard A2 milk during other periods (Figure 4).
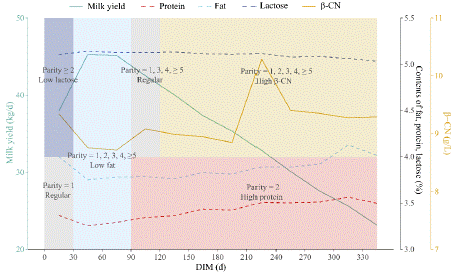
Figure 4: Strategy for A2 milk production. Attached were the trait variation
curves with DIM. The green solid line represented milk yield, with the
primary y-axis indicating its range. The red, blue, and purple dashed lines
represented protein, fat, and lactose, respectively, with the first secondary
y-axis indicating their range. The yellow solid line represented β-casein
(β-CN), with the second secondary y-axis showing its range. The purple
rectangle represented the production of low-lactose A2 milk during days
0-30 of the second and subsequent parities; the blue rectangle showed
the production of low-fat A2 milk in days 30-90 of each parity; the yellow
rectangle indicated high β-CN A2 milk production starting from day 120, and
the red rectangle highlighted high-protein A2 milk production beginning on
day 90 of the second parity. The gray rectangles corresponded to regular A2
milk production during the remaining periods.
During the various stages outlined above, the nutrient content in A2 milk was as follows: In low-lactose A2 milk, the milk protein content is relatively high (average 3.38%), with fat (4.07%), TS (13.62%), as1-casein (11.93g/L), a-La (1.01g/L), and β-LG (3.47g/L) all exceeding the stage average. In low-fat A2 milk, β-LG (3.51g/L) was higher than the average (3.29 g/L). Protein-rich A2 milk contains elevated levels of milk protein (3.46%), milk fat (3.89%), TS (13.49%), SNF (9.44%), total casein (25.08g/L), as1-CN (13.30g/L), and κ-CN (4.22g/L), surpassing average values, while lactose (5.09%) was below average. In β-CN-rich A2 milk, protein (3.46%), fat (3.89%), TS (13.46%), T-CN (25.08g/L), as1-CN (11.25g/L), κ-CN (4.25g/L), and a-LA (0.98g/L) were above average, with lower lactose content (5.09%). In regular A2 milk, the average milk protein content was 3.36%, with milk fat at 3.80%. Thus, consumption of low-lactose and low-fat A2 milk ensures intake of high-quality protein and essential amino acids, while high-protein and high-β-CN A2 milk provide higher levels of nutrients. After segregating A2 cows, the non-A2 herd still maintained high milk yield and production levels of milk fat and protein. The milk fat (3.89%) and protein content (3.39%) not only meet but also exceed the Chinese standards for food safety (fat =3.1%, protein =2.95%) [26]. Overall, the non-A2 cows were suitable as a conventional source for non-A2 milk production.
Pre-analysis of economic benefits
The online milk sale platform information showed that the price of A2 milk (about RMB 5.8 yuan/200 ml) was higher than that of conventional milk (about RMB 3.5 yuan /250 ml). Assuming a frequency of 0.30 for the A2A2 genotype within the dairy herd, based on the milk production performance of the dairy herd in this study with processing losses accounted for at 1%, processing costs at 0.35 yuan per carton and feeding costs of approximately 80 yuan per cow per day, we analyzed the economic benefits of a dairy farm with 1,000 lactating cows over 305day. All the 1,000 cows in the mixed herd produced conventional milk with an average daily milk yield of 36.04 kg/d, 305-day dairy sales of about RMB 152.35 million, and a net revenue of RMB 112.71 million after removing feeding and processing costs (approximately RMB 39.68 million). By rearing A2 cows and non-A2 cows in separate herds, the average daily milk yield of 300 A2 cows was 35.52 kg/d, and 305-day sales of A2 milk was RMB 93.32 million. The average daily milk yield of 700 non-A2 cows was 36.40 kg/d, and 305-day sales of conventional milk was RMB 107.70 million. The total milk sales were approximately RMB 201.02 million. After removing feeding and processing costs (approximately RMB 40.80 million), a net revenue of RMB 160.22 million was obtained. By separate herd rearing of A2 and non-A2 lactating cows to produce A2 milk and conventional milk, the economic benefits of farms would be improved with an extra profit of RMB 47.51 million in a lactation period (Table 2).
Milk yield,
kg/d/cowSize
305d milk yield,
tUnit,
yuan/kgIncome,
million yuanCost,
million yuanProfit,
million yuanProfit Difference,
million yuanA2 herd
35.52
300
3250.45
29
93.32
12.95
80.37
47.51
Non-A2 herd
36.40
700
7770.65
14
107.70
27.85
79.85
Sum of separated herds
1000
11021.10
201.02
40.80
160.22
Unseparated herd
36.04
1000
10991.77
14
152.35
39.63
112.71
Table 2: Pre-analysis of the economic efficiency of separate herd rearing of A2 and non-A2 cows.
The implementation of the three- division rearing strategy would not only ensure the milk yield and quality of dairy cows but also meet the needs of different consumer groups and optimize the utilization of cows as milk sources.
Conclusion
This study confirmed the patterns of milk production traits in Chinese A2 cows as varying with DIM and parity, influenced by their interaction effect. Consequently, a three -division rearing strategy was proposed based on A2 and non-A2 genotype, lactation period, and parity. This strategy would effectively enable the production of five distinct types of A2 milk: protein-rich A2 milk, high β-CN A2 milk, low-fat A2 milk, low-lactose A2 milk, and regular A2 milk. The implementation of this strategy would also enhance the economic benefits due to the improved quality of milk produced by A2 cows. After that, the remaining non-A2 cows exhibited higher milk yield, capable of generating a larger volume of milk that meets national standards. The increased milk yield of non-A2 cows consequently enhances their economic profitability.
Acknowledgments
Financial assistance from the Inter-Governmental International Science and Technology Cooperation Project of the State Key Research and Development Program (2021YFE0115500), National Center of Technology Innovation for Dairy (No. 2022-Scientific Research-3) and International Science and Technology Cooperation Project of Hubei Province (2022EHB043) are highly appreciated.
Conflicts of Interest Statement
The authors declare no real or perceived conflicts of interest.
References
- Bittante G, Penasa M, Cecchinato A. Invited review: Genetics and modeling of milk coagulation properties. Journal of Dairy Science. 2012; 95: 6843-70.
- Li C, Wang M, Cai W, Liu S, Zhou C, Yin H, et al. Genetic Analyses Confirm SNPs in HSPA8 and ERBB2 are Associated with Milk Protein Concentration in Chinese Holstein Cattle. Genes. 2019; 10.
- Amalfitano N, Stocco G, Maurmayr A, Pegolo S, Cecchinato A, Bittante G. Quantitative and qualitative detailed milk protein profiles of 6 cattle breeds: Sources of variation and contribution of protein genetic variants. Journal of Dairy Science. 2020; 103: 11190-208.
- Singh A, Kumar A, Gondro C, Pandey AK, Dutt T, Mishra BP. Genome Wide Scan to Identify Potential Genomic Regions Associated with Milk Protein and Minerals in Vrindavani Cattle. Frontiers in Veterinary Science. 2022; 9.
- Bijl E, van Valenberg HJF, Huppertz T, van Hooijdonk ACM. Protein, casein, and micellar salts in milk: current content and historical perspectives. J Dairy Sci. 2013; 96: 5455-64.
- Guantario B, Giribaldi M, Devirgiliis C, Finamore A, Colombino E, Capucchio MT, et al. A Comprehensive Evaluation of the Impact of Bovine Milk Containing Different Beta-Casein Profiles on Gut Health of Ageing Mice. Nutrients. 2020; 12.
- Hohmann LG, Yin T, Schweizer H, Giambra IJ, König S, Scholz AM. Comparative Effects of Milk Containing A1 versus A2 β-Casein on Health, Growth and β-Casomorphin-7 Level in Plasma of Neonatal Dairy Calves. Animals. 2020; 11.
- Hao W, Zhi-guo Z, Yan-zhong C, Xiang-lin D, Shu-qiang Z, Nan Z, et al. Isolation and Quantification of Main Milk Proteins in Dairy Products with Reverse-phase HPLC. Food Science. 2009; 30: 376-80.
- Caroli AM, Savino S, Bulgari O, Monti E. Detecting β-Casein Variation in Bovine Milk. Molecules. 2016; 21.
- Pal S, Woodford K, Kukuljan S, Ho S. Milk Intolerance, Beta-Casein and Lactose. Nutrients. 2015; 7: 7285-97.
- Huang M, Li H, Shao G, Wang D, Guo C, Liu W, et al. Discrimination and Breeding about A2 Beta-casein Dairy Cattle Modern Animal Husbandry Science & Technology. 2022; 11: 73-6.
- Bisutti V, Pegolo S, Giannuzzi D, Mota LFM, Vanzin A, Toscano A, et al. The β-casein (CSN2) A2 allelic variant alters milk protein profile and slightly worsens coagulation properties in Holstein cows. Journal of Dairy Science. 2022; 105: 3794-809.
- Kumar A, Singh RV, Chauhan A, Ilayakumar K, Kumar S, Kumar A, et al. Genetic association analysis reveals significant effect of β-casein A1/A2 loci on production & reproduction traits in Frieswal crossbred cows. Biological Rhythm Research. 2019; 51: 1259-72.
- Tommasoni C, Fiore E, Lisuzzo A, Gianesella M. Mastitis in Dairy Cattle: On- Farm Diagnostics and Future Perspectives. Animals (Basel). 2023; 13.
- Mk T. Effect of Nutrition on Production, Composition, Fatty acids and Nutraceutical Properties of Milk. Advances in Dairy Research. 2015; 02.
- Matamoros C, Dechow CD, Harvatine KJ. Interaction of DGAT1 polymorphism, parity, and acetate supplementation on feeding behavior, milk synthesis, and plasma metabolites. J Dairy Sci. 2023; 106: 7613-29.
- Marumo JL, Lusseau D, Speakman JR, Mackie M, Hambly C. Influence of environmental factors and parity on milk yield dynamics in barn-housed dairy cattle. J Dairy Sci. 2022; 105: 1225-41.
- Comin A, Cassandro M, Chessa S, Ojala M, Dal Zotto R, De Marchi M, et al. Effects of Composite β- and κ-Casein Genotypes on Milk Coagulation, Quality, and Yield Traits in Italian Holstein Cows. J Dairy Sci. 2008; 91: 4022- 7.
- Fatehi F, Zali A, Honarvar M, Dehghan-Banadaky M, Young AJ, Ghiasvand M, et al. Review of the relationship between milk urea nitrogen and days in milk, parity, and monthly temperature mean in Iranian Holstein cows. J Dairy Sci. 2012; 95: 5156-63.
- Sabek A, Li C, Du C, Nan L, Ni J, Elgazzar E, et al. Effects of parity and days in milk on milk composition in correlation with β-hydroxybutyrate in tropic dairy cows. Trop Anim Health Prod. 2021; 53: 270.
- Wiggans GR, Shook GE. A Lactation Measure of Somatic Cell Count.pdf. J Dairy Sci. 1987; 70: 2666–72.
- Pegolo S, Giannuzzi D, Bisutti V, Tessari R, Gelain ME, Gallo L, et al. Associations between differential somatic cell count and milk yield, quality, and technological characteristics in Holstein cows. Journal of Dairy Science. 2021; 104: 4822-36.
- Chen Y, Hostens M, Nielen M, Ehrlich J, Steeneveld W. Herd level economic comparison between the shape of the lactation curve and 305 d milk production. Frontiers in Veterinary Science. 2022; 9: 997962.
- Mohanty BS, Verma MR, Sharma VB, Patil VK. Effect of parity on the shape of lactation curves in purebred Jersey cows in Indian conditions. Biological Rhythm Research. 2019; 53: 26-39.
- Agani ZA, Pomalegni SCB, Akouedegni CG, Boko KC, Orou DB, Dossou J, et al. Ethnoveterinary study of galactogenic recipes used by ruminant breeders to improve milk production of local cows in Benin Republic. J Ethnopharmacol. 2022; 285: 114869.
- China MoHotPsRo. National food safety standard Raw milk. 2010.