
Research Article
Austin J Gastroenterol. 2023; 10(2): 1125.
The m6A Methyltransferase METTL3 Promotes Pancreatic Cancer Metastasis and Invasion
Hao Sun1; Shaolong Hao2; Yu Ji2; Xinyu Zhao3; Zhaoyu Sun4; Lang Ji1; Wei Han1,2; Yuchuan Ding5
1Central Laboratories, Beijing Luhe Hospital, Capital Medical University, Beijing, China
2Department of General Surgery, Beijing Luhe Hospital, Capital Medical University, Beijing, China
3Department of Critical Care, Beijing Lu He hospital, Capital Medical University, Beijing, China
4Medical image science, The fifth people’s hospital of Jinan, Jinan, Shandong, China
5Department of Neurosurgery, Wayne State University School of Medicine, USA
*Corresponding author: Hao Sun Central Laboratories, Beijing Luhe Hospital, Capital Medical University, Beijing, China. Email: m18810253032@163. com
Received: September 13, 2023 Accepted: October 13, 2023 Published: October 20, 2023
Abstract
Background: Pancreatic Cancer (PC) poses a formidable challenge to human health due to its high degree of malignancy, difficult diagnosis and treatment, and poor prognosis. RNA N6-methyladenosine (m6A) is a key step of posttranscriptional modulation that is involved in governing gene expression. The m6A modification catalyzed by METTL3 has been widely recognized as a critical epigenetic regulation process for tumorigenic properties in various cancer cell lines, including pancreatic cancer, but its specific mechanism has remained elusive.
Methods: Transcriptional profiles and clinical data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) were analyzed. Through Gene Set Enrichment Snalysis (GSEA) analysis, Western blot, and qPCR, we confirmed the high expression of METTL3 and the increased m6A methylation level in tumor tissues. And METTL3 high expression decreased survival. We constructed stable METTL3 knockout cell lines and found that METTL3 knockout inhibited tumor migration and invasion.
Results: According to the analysis of the TCGA clinical database, METTL3 exhibits higher expression in tumor tissues compared to normal tissues. METTL3 expression was higher in pancreatic cancer tumor tissues compared to paracancerous tissue. Gene Set Enrichment Analysis (GSEA) revealed that METTL3 promotes pancreatic cancer progression, potentially through its role in methylation. Western blot and qPCR experiments confirmed these findings. Constructing stable METTL3 knockout cell lines revealed that METTL3 knockout inhibited tumor migration and invasion. Furthermore, our results confirmed that METTL3 impacts the malignant progression of pancreatic cancer via the VEGF/VEGFR pathway.
Keywords: PDCA; METTL3; m6A; Metastasis; Invasion
Introduction
Pancreatic cancer is a malignancy of the digestive tract that exhibits insidious and atypical clinical symptoms, rendering it challenging to diagnose and treat effectively. Around 90% of cases are Pancreatic Ductal Adenocarcinoma (PDAC) which ductal adenocarcinomas arising in the glandular duct epithelium. The development of pancreatic cancer is closely linked to lifestyle factors such as smoking, drinking, and diet. Unfortunately, early diagnosis of pancreatic cancer is uncommon, with high mortality rates associated with surgical intervention and low rates of complete remission [1,2]. Notably, pancreatic cancer boasts the lowest 5-year survival rate of all tumors (less than 8%) [3,4]. As such, early diagnosis and prompt intervention for patients with pancreatic cancer are essential to optimize treatment outcomes.
N6-Methyladenosine (m6A) is the most common methylation modification that occurs in eukaryotic mRNA sequences following transcription. It is enriched in the consensus RRACH motif located near the 3' non-coding region, the 5' non-coding region, and the stop codon. (R represents A or G, while H denotes A, C, or U). The process of m6A methylation is highly dynamic and involves three key types of enzymes: Writers, Erasers, and Readers [5,6]. methyltransferases are responsible for catalyzing adenylate to modify m6A on mRNA while demethylases work to remove the m6A modified base. Reading proteins, on the other hand, are responsible for identifying the base of m6A modification and activating downstream regulatory pathways. Through synergistic action with downstream recognition proteins, regulation of mRNA splicing, nucleation, localization, translation, and stability can be achieved [5,7], The functional regulation of transcriptional and post-transcriptional processes in eukaryotic cells is therefore achieved, with significant effects on stress response, cell reproduction, stem cell differentiation, development, and tumor occurrence [8,9].
METTL3 as a crucial component of the m6A methylated transferase complex, is a methyltransferase involved in the process of RNA modification through the addition of m6A. It forms stable heterodimers with methyltransferase 14(METTL4) with Wilm's Tumor 1-Associated Protein (WTAP) as a cofactor, thus constituting the main structure of m6A methylated transferase [10,11]. METTL3, also referred to as MT-A70, has two critical domains that bind S-Adenosylmethionine (SAM) and catalyze the formation of m6A, respectively [12,13]. Additionally, both METTL3 and METTL14 are core proteins of m6A methyltransferase complex [14]. Research has shown that METTL3 can impact the progression of various types of cancers such as gastric cancer [15,16], lung cancer [17], pancreatic cancer [18] and other tumors by affecting RNA methylation modification. . However, the mechanism by which METTL3 promotes malignant progression of pancreatic cancer remains understudied, making our results important for early diagnosis of pancreatic cancer and providing targeted therapy for METTL3.
Methods
Human Specimens
The study was performed after approval by the Institutional Ethical Board of our affiliated hospital. All enrolled individuals provided informed consent. Paired distant normal tissues away from the tumor margins at least 5 cm were served as controls.
Cell Lines and Transfection
PANC-1, SW1990 were were purchased from Guangzhou Genio Biotech Co., Ltd. The SW1990 and PANC-1 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Millipore Sigma) supplemented with 10% heat-inactivated fetal bovine serum FBS (Thermo Fisher Scientific, Inc.) and 1% antibiotics (penicillin-streptomycin; Thermo Fisher Scientific, Inc.). All cells were performed in a humidified incubator at 37°C and in a moist incubator stabilized at 5% CO2. To knock down endogenous METTL3. Lipofectamine™ 3000 (Thermo Fisher Scientific, Inc.) was used to transfect the PC cell lines with a small interfering siMETTL3 [19].
RNA Extraction and Quantitative Real-Time PCR
A TRIzol® RNA extraction kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used for isolation of total RNA from cell and tissue according to the manufacturer's instruction. Subsequently, we used a First-Strand cDNA Synthesis Kit ((Takara Biotechnology Co., Ltd.) to obtain the complementary DNA (cDNA). For real-time PCR (qRT-PCR), amplifications were performed by the real-time PCR Master Mix (SYBR® Primx Ex Taq™ (TIi RNaseH Plus) (cat. no. RR420A; Takara Bio, Inc.). We used 2 - ΔΔCt method to analyze the mRNA expression. GAPDH expression was used as internal controls. The primer sequence of the genes was shown is shown in Table 1.
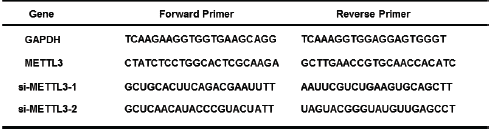
Table 1: Primer and siRNA sequences.
Western Blotting
All tissue/cell line samples wereapplied to extract protein by using RIPA buffer r (cat. no. R0010; Beijing Solarbio Science & Technology Co., Ltd.) and protein was quantified using the bicinchoninic acid (BCA) method (Thermo Fisher Scientific, Inc.). The remaining specific steps of Western Blot (WB) refer to reported previously [20].
Wound-Healing Assay
After transfection, PC cells were seeded onto 6-well plates and a confluent monolayer was achieved once the cells reached 80% confluences. The floating cells were removed by washing with 1×PBS. Cell migration was monitored and captured at different time points using a phase contrast microscope and ImageJ software to measure the area of migration into the wound and the width of the healed wound. Changes in wound width were measured linearly from the initial wound width at 0 h to observe cellular migratory ability. The data generated from this experiment were used to assess cell motility.
Transwell Migration Assay and Matrigel Invasion Assay
Cell migration and invasion assays were conducted using 8μM Transwell chambers (Catalog No. 3422, Corning, USA). For the migration experiment, cells were seeded into the top chamber in serum-free medium, while the lower chamber contained 0. 5 mL of medium with 10% FBS. Following incubation at 37°C in a 5% CO2 environment for 24 h, the upper side of the membrane was carefully wiped with a cotton swab to remove the non-migrating cells, while the migrating cells were stained with crystal violet and counted.
For the invasion experiment, the top side of the Transwell membrane was first coated with Matrigel and incubated for 60 minutes at 37°C. The cells starved for 24 hours and 200μL suspension containing 5×104 SW1990 or PANC-1 cells was added to the top chamber, and 0. 5 mL of medium with 10% FBS was added to the bottom chamber. The cells were then incubated under the same conditions as the migration assay. After 24 h, non-invasive cells were removed from the uppermost surface of the membrane, and invasive or migrated cells were fixed with 4% paraformaldehyde and stained with 2% crystal violet. The invasive or migrated cells were counted and the data were analyzed.
m6A RNA Methylation Quantifcation
The EpiQuik m6A Methylation Quantitative Kit (colorimetric; Epigentek) was utilized to determine the overall m6A level present in the extracted RNA, in accordance with the manufacturer's instructions [21]. For each individual sample analysis, a quantity of 200 ng poly-apurified RNA was employed.
Functional Enrichment Analysis
To perform correlation and differential expression analyses of the m6A family, RNAseq data from TCGA PAAD were obtained from the UCSC Xena website (https://xenabrowser. net/). In addition, expression profiles of PC tissues and adjacent normal tissues were obtained from an array and downloaded from the GEO database (GEO: GSE15471) [18]. The Broad GSEA software (version 4. 0. 2) was utilized to perform GSEA, employing the gene sets within MSigDB for pathway annotation. Using the annotated gene set from the MSigDB database, we conducted GSEA to identify variations in corresponding pathways between different risk groups (P<0. 05, False Discovery Rate (FDR) <0. 25) [19].
Result
METTL3 Over Expression Correlates with Poor Prognosis of Pancreatic Cancer Patients
First we compared the RNA-seq data downloaded from GEO (GDS4103) and TCGA databases mRNA expression levels of METTL3 from GEO, and we found that the mRNA expression levels of METTL3 were upregulated in tumor samples compared with them in paracancerous tissues (Figure 1A, B). Mettl3 had a negative impact on the overall survival time of patients with pancreatic cancer (Figure 1C). In addition, GSEA analysis showed that the enrichment of highly expressed genes in pancreatic cancer affected the malignant progression of PDAC in cases with high METTL3 expression (Figure 1D). Promoting the enrichment of M6A methylation-related genes in pancreatic cancer patients with high METTL3 expression; promoting the enrichment of genes associated with demethylation modification in pancreatic cancer patients with low METTL3 expression (Figure 1E). These results suggest that high expression of METTL3 in PC tissues correlates with a poor prognosis. This is also the same as the literature results [18]. We measured m6A levels in tissues.
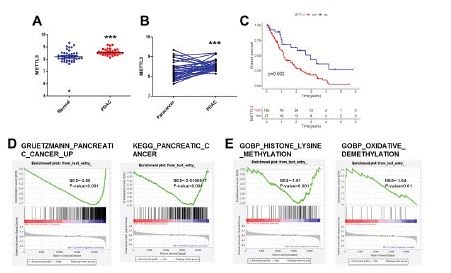
Figure 1: METTL3 is highly expressed in PC tissues and is associated with poor prognosis in pancreatic cancer patients. (A) Comparisons of METTL3 in normal and PDAC tissues. (B) Comparisons of METTL3 in paracancer and cancerous tissue. (C) The OS KM curves between low- and high-METTL3 groups in the GEO(GDS4103) cohort. (D) The GSEA of the PC gene set in low- and high-METTL3 groups. (E) The GSEA of the methylation and demethylation gene set in low- and high-METTL3 groups ***p<0.001.
The Level of m6A Increased in PC Tissues
Through GSEA analysis, we found that high expression of METTL3 can increase the methylation level and reduce demethylation in tumor tissues in patients with pancreatic cancer. We collected tumor tissue from 4 pancreatic cancer patients and compared the mRNA and protein expression levels of METTL3 between pancreatic tumor tissues and paracancerous tissues and found that the mRNA and protein expression levels of METTL3 were upregulated in tumor samples compared with them in paracancerous tissues (Figure 2A, B). Mettle3 is the core protein that catalyzes the formation of m6A.
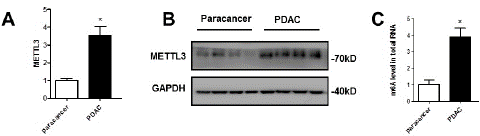
Figure 2: The level of m6A increased in PC tissues. (A)(B) Comparisons of METTL3 in paracancer and cancerous tissue. (C) m6A methylation levels of PDAC were analyzed ***p<0.05.
Mettl3 can affect the malignant progression of tumors through m6A. we measured m6A levels in PC tissue, the m6A levels were upregulated in tumor samples compared with them in paracancerous tissues (Figure 2C). The results showed that the level of m6A in pancreatic cancer tissue was significantly higher than that in the adjacent tissues.
METTL3 Knockout Reduces the m6A Level in PC
METTL3 is highly expressed in PC tissues and also in pancreatic cancer cell lines. Through the THPA database, we found that METTL3 was highly expressed in pancreatic cancer cell lines (Figure 3A). PC cell lines panc-1 and sw1990 were used as cell models. We constructed the METTL3 stable knockout cell lines panc-1 and sw1990. The expression of METTL3 in cells was significantly decreased (Figure 3B, C).
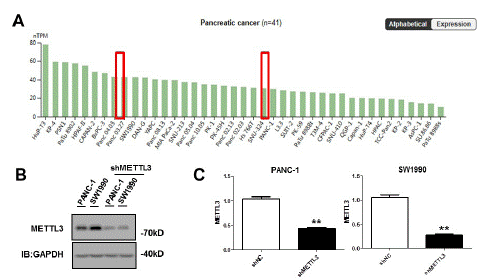
Figure 3: METTL3 knockout reduces the m6A level in PC. (A) METTL3 is highly expressed in THPA in pancreatic cancer cell lines. (B) (C) Construct stable METTL3 knockdown cell lines in PANC-1 and SW1990 cell lines. The expression of METTL3 protein was detected by Western blot and qPCR **p<0.01.
METTL3 Knockout Inhibited Migration and Invasion of PC Cells
Gene Set Enrichment Analysis (GSEA) was performed using the GEO public database (GSE183795). Genes positively associated with tumor metastasis were found to be enriched in pancreatic cancer patients with high METTL3 expression (Figure 4A). Moreover, abnormal gene expression during lymphatic infiltration was enriched in patients with high METTL3 expression (Figure 4B, C). The effect of shMETTL3 on the progression of pancreatic cancer was found by cell function experiments. METTL3 knockout decreased the migration and invasion ability of pancreatic cancer cells by scratch assay and Transwell assay (Figure 3D, E). Therefore, we have confirmed that METTL3 plays a role in promoting cancer in pancreatic cancer, high expression of METTL3 can increase the proliferation, migration and invasion ability of pancreatic cancer and reduced expression of METTL3 can reduce the malignant progression of tumor.
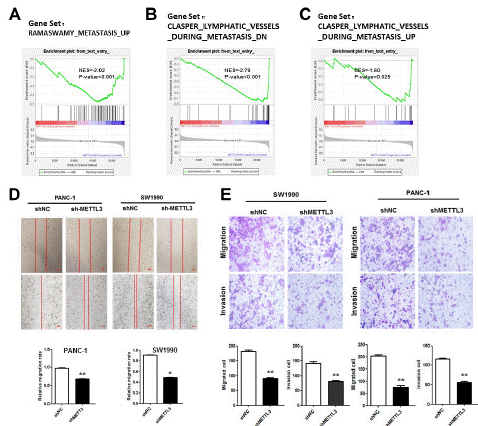
Figure 4: Abnormally high expression of METTL3 promotes metastasis of pancreatic cancer.
(A) The GSEA of the METASTASIS gene set in low- and high-METTL3 groups. (B) The GSEA of the CLASPER LYMPHATIC VESSELS DURING METASTASIS DN gene set in low- and high-METTL3 groups. (C) The GSEA of the CLASPER LYMPHATIC VESSELS DURING METASTASIS UP gene set in low- and high-METTL3 groups. (D) Wound-healing analysis of the cell migrative potential after the transfection with siMETTL3 treatment in SW1990 and PANC-1 cell lines (scar bar: 50 μm). (E) Transwell analysis of the cell invasive potential after the transfection with siMETTL3 treatment in SW1990 and PANC-1 cell lines *p<0.05; **p<0.01.
METTL3 Promotes Pancreatic Cancer Metastasis through the VEGF/VEGFR Pathway
Through the above experiments, we concluded that METTL3 could promote the metastasis and invasion of pancreatic cancer (Figure 4D, E). GSEA found that its migration and invasion ability was correlated with lymph node metastasis, so we analyzed the VEGF/VEGFR pathway closely related to tumor metastasis. Activation of the pathway was found to be significantly enriched in cases with high METTL3 expression (Figure 5A). Further, we analyzed the correlation between METTL3 and VEGF, and there was a significant positive correlation between METTL3 and VEGFA/VEFGB/VEGFC (Figure 5B).
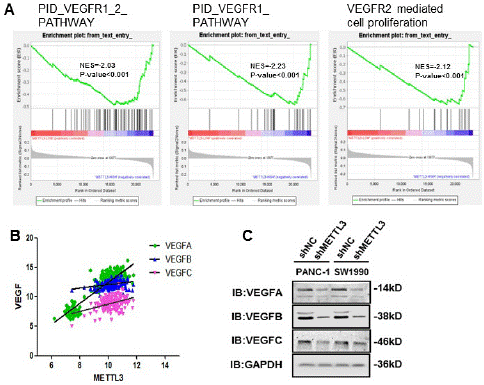
Figure 5: METTL3 promotes pancreatic cancer metastasis through the VEGF/VEGFR pathway. (A) The GSEA of the VEGFR1_2_PATHWAY gene set in low and high-METTL3 groups. (B) Correlation analysis of METTL3 and VEGF (VEGFA: Spearman r=0.6917, p<0.0001; VEGFB: Spearman r= 0.3090, p<0.0001; VEGFC: Spearman r= 0.2793, p<0.0001). It was analyzed according to the TCGA database. (C) VEGF family proteins were compared in stable METTL3 knockdown cell lines.
We found that knockdown of METTL3 in PANC-1 and SW1990 cell lines decreased protein levels of VEGFA/B/C (Figure 5C). VEGFA/VEFGB mainly affects vascular metastasis of tumors, while VEGFC affects lymph node metastasis of tumors. This suggests that METTL3 can promote vascular and lymphatic metastasis of pancreatic cancer through VEGF/VEGFR [22].
Discussion
Pancreatic cancer is the most malignant tumor with strong metastasis and poor prognosis, which seriously affects human life and health. Therefore, the discovery of biomarkers and mechanism of early pancreatic cancer is the focus of pancreatic cancer treatment. As a key component of m6A methylated transferase complex, METTL3 has been reported to be highly expressed in pancreatic cancer tissues, which is correlated with the progression and prognosis of PC [18]. The m6A modification catalyzed by METTL3 control specific target mRNA translation and stability, which has been proven to promote progression of various cancer including leukemia, endometrial cancer, lung cancer and colorectal cancer [23-26]. A previous study has shown that METTL3 mediates m6A-TEK-VEGF-A passway to promote tumor angiogenesis in bladder cancer [27]. However, research on the role of METTL3 in PC is limited, and currently there are studies that have found METTL3 to be overexpressed in pancreatic cancer and associated with poor prognosis, but the underlying mechanisms promoting cancer remain unknown.
Our findings demonstrate that METTL3 is highly expressed in pancreatic cancer tissues, showing differential expression between tumor and adjacent normal tissues. Our analysis of public databases (TCGA and GEO) and pathological tissue samples shows that METTL3 overexpression is associated with poor prognosis. Furthermore, GSEA analysis revealed that the METTL3 high-expression group exhibited increased methylation modification, decreased demethylation, and significant enrichment of oncogenic genes. Importantly, we observed a significant correlation between high METTL3 expression and tumor metastasis and invasion in pancreatic cancer. In vitro experiments also confirmed that knocking down METTL3 significantly reduced the migration and invasion ability of PC cell lines PANC-1 and SW1990. These findings may contribute to early diagnosis of pancreatic cancer and provide a targeted therapy approach by targeting METTL3.
Therefore, we analyzed the VEGF/VEGFR pathway, which is closely related to tumor metastasis and invasion. The activation of the VEGF/VEGFR pathway was also enriched in the METTL3 high expression group, which is associated with pancreatic cancer vascular and lymph node metastases. The VEGF (vascular endothelial growth factor) family includes multiple members, primarily VEGFA, VEGFB, and VEGFC. VEGFA is the most significant member of the VEGF family and can promote endothelial cell proliferation, migration, and tube formation, thereby promoting angiogenesis. VEGFB has relatively complex functions, including promoting new blood vessel generation, inducing cell apoptosis, and enhancing vascular permeability. VEGFC mainly promotes lymphangiogenesis and lymphatic expansion by activating VEGFR-3. Our findings suggest that METTL3 expression positively correlates with VEGF expression, especially VEGFA. Targeting VEGFA has the potential to serve as an anti-tumor therapy for pancreatic cancer, and our study provides a new candidate target for VEGFA-based therapy in pancreatic cancer.
Our results demonstrate a significant positive correlation between METTL3 and VEGF expression, particularly with VEGFA which is the most common target of anti-tumor targeted drugs. Therefore, understanding the mechanism of METTL3 in disease is crucial for the clinical diagnosis and targeted therapy of pancreatic cancer, particularly in relation to VEGFA-based therapy.
Author Statements
Author Contributions
Writing—original draft preparation, H.S.and S.H., and they contributed equally to this work.Conceptualization, H.S., L.H., S.H., Y.D.Y; methodology, H.S., Y.J., S.H., W.H., Y.D.; software, X.Z.,L.J.; validation, H.S., S.H., Y.J., Z.S., L.J., W.H,.Y.D.; formal analysis, H.S., S.H., X.Z.; investigation, H.S., Z.S., Y.J., S.H., W.H,.Y.D.; data curation, H.S., S.H..; writing—review and editing, H.S., S.H., Y.J., Y.D.; visualization, H.S., Z.S., X.Z., W.H., Y.D.; supervision, H.S., S.H., W.H., Y.D.; project administration, H.S., W.H., Y.D.; funding acquisition, N/A.All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the Science and Technology Plan of Beijing Tongzhou District (KJ2022CX016) and Beijing Municipal Natural Science Foundation (7234377).
Availability of Data and Materials
All the data generated or analyzed during this study are included in this published article and its Additional files.
Conflicts of Interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Consent for Publication
All the authors read and approved the final manuscript.
Competing Interests
The authors declare no potential competing interests.
References
- Halbrook CJ, Lyssiotis CA, Pasca di Magliano M, Maitra A. Pancreatic cancer: advances and challenges. Cell. 2023; 186: 1729-54.
- Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD, et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature. 2023; 615: 168-74.
- Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020; 395: 2008-20.
- Chari ST, Sharma A, Maitra A. Early detection of sporadic pancreatic ductal adenocarcinoma: problems, promise, and prospects. Ann Intern Med. 2020; 172: 558-9.
- Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The critical role of RNA mA methylation in cancer. Cancer Res. 2019; 79: 1285-92.
- Flamand MN, Tegowski M, Meyer KD. The proteins of mRNA modification: writers, readers, and erasers. Annu Rev Biochem. 2023; 92: 145-73.
- Pan Y, Ma P, Liu Y, Li W, Shu Y. Multiple functions of mA RNA methylation in cancer. J Hematol Oncol. 2018; 11: 48.
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014; 15: 293-306.
- Wu L, Wu D, Ning J, Liu W, Zhang D. Changes of N6-methyladenosine modulators promote breast cancer progression. BMC Cancer. 2019; 19: 326.
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24: 177-89.
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014; 10: 93-5.
- Leach RA, Tuck MT. Expression of the mRNA (N6-adenosine)-methyltransferase S-adenosyl-L-methionine binding subunit mRNA in cultured cells. Int J Biochem Cell Biol. 2001; 33: 984-99.
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA (N Y NY). 1997; 3: 1233-47.
- Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016; 534: 575-8.
- Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020; 521: 887-93.
- Liu T, Yang S, Sui J, Xu SY, Cheng YP, Shen B, et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020; 235: 548-62.
- Wei W, Huo B, Shi X. miR-600 inhibits lung cancer via downregulating the expression of METTL3. Cancer Manag Res. 2019; 11: 1177-87.
- Li Y, Huang H, Zhu Y, Xu B, Chen J, Liu Y, et al. Increased expression of METTL3 in pancreatic cancer tissues associates with poor survival of the patients. World J Surg Oncol. 2022; 20: 283.
- Zhou Y, Jiang R, Jiang Y, Fu Y, Manafhan Y, Zhu J, et al. Exploration of N6-methyladenosine profiles of mRNAs and the function of METTL3 in atherosclerosis. Cells. 2022; 11: 2980.
- Hao S, Han W, Ji Y, Sun H, Shi H, Ma J, et al. BANCR positively regulates the HIF-1a/VEGF-C/VEGFR-3 pathway in a hypoxic microenvironment to promote lymphangiogenesis in pancreatic cancer cells. Oncol Lett. 2022; 24: 422.
- Peng J, Zheng H, Liu F, Wu Q, Liu S. The m6A methyltransferase METTL3 affects autophagy and progression of nasopharyngeal carcinoma by regulating the stability of lncRNA ZFAS1. Infect Agents Cancer. 2022; 17: 1.
- Patel SA, Nilsson MB, Le X, Cascone T, Jain RK, Heymach JV. Molecular mechanisms and future implications of VEGF/VEGFR in cancer therapy. Clin Cancer Res. 2023; 29: 30-9.
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millán-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by mA-dependent translation control. Nature. 2017; 552: 126-31.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68: 394-424.
- Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018; 561: 556-60.
- Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, et al. The mA methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019; 38: 3667-80.
- Wang G, Dai Y, Li K, Cheng M, Xiong G, Wang X, et al. Deficiency of Mettl3 in bladder cancer stem cells inhibits bladder cancer progression and angiogenesis. Front Cell Dev Biol. 2021; 9: 627706.