
Special Article - Digestive Endoscopy
Austin J Gastroenterol. 2015;2(1): 1031.
Determination of Helicobacter pylori Virulence Genes in Clinical Isolates of Symptomatic Patients from South Coastal Region of Karnataka - A Preliminary Work
Shetty V1, Ballal M1*, Lingadakai R2 and Mukhopadhyay AK3
1Department of Microbiology, Manipal University, India
2Department of Surgery, Manipal University, India
3Bacteriology Division, National Institute of Cholera and Enteric Diseases, Kolkata
*Corresponding author: Ballal M, Department of Microbiology, Enteric Pathogens Division, Manipal University, Manipal, Karnataka State, Pin: 576104, India
Received: November 28, 2014; Accepted: January 28, 2015; Published: January 30, 2015
Abstract
Introduction: Helicobacter pylori is a microaerophilic, spiral shaped, unipolar flagellated, gram negative organism which has high tropism towards gastric epithelial cells and may lead to a severe form of gastroduodenal disease. Prevalence of H pylori infections in developing countries may exceed 70% but only a fraction of people develop severe disease. The involvement of H. pylori virulence factors cagA and vacA genotypes are well studied and show high genetic diversity geographically. The present study was conducted to detect the Presence of H. pylori in biopsy samples from symptomatic patients by Rapid Urease Test, histopathological examination and Culture .It also involved the detection of virulence genes cagA, vacA (s1m1, s1m2 and s2m2) genotypes from the isolated strains.
Materials and Methods: Patients with complaints of abdominal pain, discomfort, acidity and loss of appetite were chosen for endoscopy. A detailed history was taken and physical examination of the patients was carried out prior to endoscopy. A total of 3 antral biopsies were obtained from each patient and subjected to RUT, HPE and culture. Isolated H. pylori were further examined for the virulence genes by using PCR.
Results: Antral biopsies were collected from a total of 38 patients. Identification of H. pylori was performed on all gastric biopsies by RUT, HPE and Culture. The rate of positive H. pylori in the biopsies tested with RUT were (36.9%), HPE (42.1%) and Culture (39.5%) and the presence of virulence gene cagA (93.3%) and vacA alleles- s1m1 (60%), s1m2 (40%). None of them showed presence of vacA s2m2 genotypes by PCR.
Conclusion: This study provides important information regarding the rate of identification of Helicobacter pylori from RUT, HPE, Culture and also rate of virulence genes harbored by strains isolated from this part of Karnataka., India.
Keywords: H. pylori; cagA gene; vacA gene; PCR; Biopsy
Introduction
Helicobacter pylori is a slow-growing, spiral shaped, 2 to 6 unipolar flagellated, gram negative microaerophilic organism which inhabits the human stomach and has a high tropism for the gastric epithelium causing long term colonization of the gastric mucosa. It has been linked to primary cause of upper gastrointestinal disorders, such as chronic active gastritis, peptic ulcer, gastric cancer and MALT lymphoma [1-3]. Most importantly it is a significant risk factor for developing gastric cancer and WHO has designated this bacterium as class 1 carcinogen [4]. The prevalence of H. pylori infection varies geographically, and it may exceed 70% in developing countries. The prevalence of H. pylori infection is high (49.94%-83.30%) in India, interestingly, only fraction of people get infection in their lifetime [5- 8].
There are several factors which are involved in severity of infection like environmental, host genetic and bacterial virulence factors [9]. The involvement of H. pylori virulence factors like cytotoxin associated gene (cagA) and vacuolating cytotoxin gene (vacA) genotypes are well studied and showed the high genetic diversity geographically. The cagA gene, a marker for the cag Pathogenicity Island (cagPAI) which includes a number of other genes associated with increased virulence, is associated with severe clinical disease and vacA gene encodes for the vacuolating cytotoxin, the pore forming toxin which leads to gastric epithelial cell injury. The vacuolating activity in host cells varies due to mosaicism of the vacA gene in signal (s) and median (m) regions [10-12].
The aim of the present study was to detect H. pylori from gastric biopsy of symptomatic patients and comparison of different methods used for their detection (RUT, HPE, and Culture) and also to determine the rate of virulence genes, cagA and vacA and their alleles by PCR in this part of south costal region of Karnataka, India.
Methods and Materials
Specimen collection and Processing
A total of 38 adult patients of both genders having complaints of abdominal pain, discomfort, acidity and loss of appetite were chosen for endoscopy and detailed history was taken and a physical examination was carried out prior to endoscopy. Patients’ consent to participate in the study was obtained as per the protocol of institutional ethical committee. A total of 3 Antral biopsies were obtained from each patient and one subjected to rapid urease test, one in 10% formalin processed for histopathological examination and one in 0.6ml brucella broth with 15% glycerol as transport media stored at -70° C until cultured.
Histopathological examination
Tissue biopsies treated with formalin was stained routinely with Haematoxylin and Eosin, special stains such as Giemsa was used as and when required.
H. pylori Culture
Transport medium containing the biopsy samples were vortexed, and later 100 μl plated on BHIA supplemented with, 0.4% IsovitaleX, 7% horse serum, and H. pylori dent supplement. Cultured plates then incubated in a microaerobic atmosphere (5% O2, 10% CO2, and 85% N2) at 37°C for a period of 3 to 7 days. H. pylori identified based on their typical morphology, gram staining, and positive reaction for urease, oxidase and catalase test.
Extraction of genomic DNA
Cells were harvested from culture plates and washed with PBS (pH8.0). The CTAB (hexadecyltrimethyl ammonium bromide) method was used for extracting the DNA.
PCR amplification
PCR amplification was done in final volume of 20μl which contained 2μl template DNA, 2μl dNTP, 1.2μl Mgcl2, 2μl 10x buffer, and10 pmol of the appropriate primers (Table 1) in the presence of 1U Taq DNA Polymerase. The cycling condition were: Initial denaturation at 95° C for 3 min, followed by 30 cycles of denaturation at 94° C for 30sec, annealing at 55° C for 30 sec and extension at 72° C for 1 min, with final extension at 72° C for 7 min.
Primers
(5’-3’) Sequence
(bp) Size
Anneling temp. ºC
cagA5cf
cagA3cR
GTTGATAACGCTGTCGCTTCA
GGGTTGTATGATATTTTCCATAA
350
55
Va1F
Va1R
ATGGAAATACAACAAACACAC
CTGCTTGAATGCGCCAAAC
259(S1)
287(S2)
55
vagF
vagR
CAATCTGTCCAATCAAGCGAG
GCGTCAAAATAATTCCAAGG
550(m1)
600(m2)
55
Table 1: Primer used for amplification of H. pylori genes.
Results
A total of 38 dyspeptic patients, 25 males and 13 Females ranging in age from 22 to 72 years (average age of 49.6 + 13.3) were included in the study. Antral biopsies were collected from 38 patients. Identification of H. pylori was performed on all gastric biopsies by Rapid Urease test, Histopathology and Culture. The rate of positive H. pylori in the biopsies tested with RUT were (36.9%), HPE (42.1%) and Culture (39.5%) (Table 2). Histopathology of the samples positive for H. pylori revealed, mild and chronic active gastritis (n=11), follicular gastritis (n=3) and chronic gastritis with Intestinal metaplasia (n=2).
RUT
HPE
Culture
Positive
14(36.9%)
16(42.1%)
15(39.5%)
Negative
24(63.1%)
22(57.9%)
23(60.5%)
Total
38(100%)
38(100%)
38(100%)
Table 2: Rate of detection of H. pylori.
The distribution of cagA gene and vacA gene (found in different allelic forms depending on the genotypes of the signal region and middle region) among 15 isolated strains was determined using specific primers (Figure 1) and the presence of virulence gene cagA (93.3%) and vacA alleles- s1m1 (60%), s1m2 (40%), no vacA s2m2 genotype from isolated H. pylori strains were determined by PCR. Virulence genes combinations show that 8 H. pylori strains had the vacAs1m1/cagA+ combination, while 1 were vacAs1m1/cagA- and 6 were vacAs1m2/cagA+ genotypes observed (Table 3).
Gene (N=15)
cagA
Positive
Negative
vacA
s1m1
8 (53.3%)
1(6.7%)
s1m2
6(40%)
-
s2m2
-
-
Table 3: Distribution of cagA and vacA genotypes.
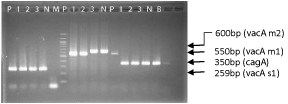
Figure 1: PCR products from clinical strains showing vacA gene [allele s1(259-bp) and m1(550-bp), m2(600-bp)] and cagA gene(350-bp). Lane PPositive control, Lane N- Negative control, Lane B- Blank, Lane M, 100-bp molecular size marker.
Discussion
A reliable primary diagnosis of H. pylori infection is crucial in management of infection for patients with different patterns of disease. This study was conducted to compare the results of rate of identification of H. pylori infection from invasive methods by three different tests like Rapid Urease Test (RUT), Histopathological Examination (HPE) and culture. The rate of positivity of H. pylori in the biopsies by HPE, culture and RUT were 42.1%, 39.5% and 36.9% respectively.
RUT was easy to perform, less expensive and more rapid when compared to histopathology and culture. The rate of positivity for H. pylori is more by HPE than Culture and RUT and this may be due to some factors like delay in transportation of samples to the laboratory, organism not being present at the region where the biopsy was taken or exposure of biopsy sample to aerobic condition that resulted in H. pylori not growing in culture plate. The lower percentage of positivity of RUT in comparison with HPE and culture may be due to changes in pH of the gastric mucosa that would have led to less sensitivity of the RUT [13].
Studies on H. pylori have shown that there are two important virulence factors - vacA and cagA genes. Both genes are involved in production of two distinct toxins which is able to affect cell shape and immune cells and may involve in severe gastric disease [14]. The amount of toxins produced corresponds with the several alleles of vacA genotypes where s1m1 produces more toxin, s1m2 produces moderate toxin and s2m2 produces few or no toxin. Heterogeneity of vacA alleles may be an important factor in understanding variations in clinical manifestations [15]. Rate of virulence genes harbored by strains isolated from this part of Karnataka showed 90% positivity for cagA genes. The polymorphic nature of vacA alleles were also studied and were found to be s1m1 (60%), s1m2 (40%) and no strains harboring s2m2 genotypes.
Helicobacter pylori, is known to harbor virulence genes which are implicated in gastric epithelial damage including gastric mucosal atrophy. Though it exists as a colonizing flora in some proportion of the individuals, it is essential to screen and identify those harboring the virulent pathotypes so that their progression to gastric and related carcinomas can be prevented. This study provides important information regarding the rate of identification of H. pylori using RUT, HPE, Culture and also their comparison and rate of virulence genes harbored by strains isolated from this part of Karnataka. Since the present work is a preliminary work with small sample size we were unable to correlate the virulence factor with disease pattern. Further analysis of strain diversity and its association with clinical disease is needed to clarify the pathogenicity of H. pylori. The above work is first of its kind on Helicobacter from this part of the country.
References
- Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006; 19: 449-490.
- Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008; 134: 306-323.
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984; 1: 1311-1315.
- [No authors listed]. An international association between Helicobacter pylori infection and gastric cancer. The EUROGAST Study Group. Lancet. 1993; 341: 1359-1362.
- Lindkvist P, Asrat D, Nilsson I, Tsega E, Olsson GL, Wretlind B, Giesecke J. Age at acquisition of Helicobacter pylori infection: comparison of a high and a low prevalence country. Scand J Infect Dis. 1996; 28: 181-184.
- Sitas F, Yarnell J, Forman D. Helicobacter pylori infection rates in relation to age and social class in a population of Welsh men. Gut. 1992; 33:1582.
- World Gastroenterology Organisation Global Guidelines. Helicobacter pylori in developing countries August 2010.
- Misra V, Pandey R, Misra SP, Dwivedi M1. Helicobacter pylori and gastric cancer: Indian enigma. World J Gastroenterol. 2014; 20: 1503-1509.
- Zabaleta J1. Multifactorial etiology of gastric cancer. Methods Mol Biol. 2012; 863: 411-435.
- Chattopadhyay S, Datta S, Chowdhury A, Chowdhury S, Mukhopadhyay AK, Rajendran K, et al. Virulence genes in Helicobacter pylori strains from West Bengal residents with overt H. pylori-associated disease and healthy volunteers. J Clin Microbiol. 2002; 40: 2622- 2625.
- Chattopadhyay S, Patra R, Ramamurthy T, Chowdhury A, Santra A, Dhali GK, B et al. Multiplex PCR assay for rapid detection and genotyping of Helicobacter pylori directly from biopsy specimens. J Clin Microbiol. 2004; 42: 2821-2824.
- Essawi T, Hammoudeh W, Sabri I, Sweidan W, Farraj MA. Determination of Helicobacter pylori Virulence Genes in Gastric Biopsies by PCR. ISRN Gastroenterol. 2013; 2013: 606258.
- Ricci C, Holton J, Vaira D. Diagnosis of Helicobacter pylori: invasive and non-invasive tests. Best Pract Res Clin Gastroenterol. 2007; 21: 299-313.
- Marc Roger Couturier. The Evolving Challenges of Helicobacter pylori Disease, Diagnostics, and Treatment, Part 1. Clinical Microbiology Newsletter. 2013; 35 : 19-24.
- Jones KR, Whitmire JM, Merrell DS. A Tale of Two Toxins: Helicobacter Pylori CagA and VacA Modulate Host Pathways that Impact Disease. Front Microbiol. 2010; 1: 115.