
Review Article
Austin J Gastroenterol. 2016; 3(1): 1059.
Microbial Biofilms, Colorectal Inflammation and Cancer
Chen Y, Peng Y* and Fu X*
Department of Gastroenterology, Affiliated Hospital of Southwest Medical University, China
*Corresponding author: Yan Peng, Department of Gastroenterology, Affiliated Hospital of Southwest Medical University, Street Taiping 25#, Region Jiangyang, Sichuan, China
Xiangsheng Fu, Department of Gastroenterology, Affiliated Hospital of Southwest Medical University, Street Taiping 25#, Region Jiangyang, Sichuan, China
Received: March 17, 2016; Accepted: May 10, 2016; Published: May 12, 2016
Abstract
The human gastrointestinal tract is colonized throughout its length by complex luminal and mucosal microbiota. Recently, there has been an upsurge in interest in the role of microbial communities that occur in biofilms on surfaces in the gut. Owing to their proximity to host tissues, mucosal bacteria interact more readily with the gut epithelium and immune system than their luminal counterparts, and recent researches indicate that they play an important role in the pathogenesis of the colorectal tumors. In this review, we will illustrate the association between microbial biofilms and epithelial cells, inflammation and carcinogenesis of the colorectum. Progress in the management of microbial biofilms is also illustrated.
Keywords: Biofilms; Epithelial cells; Colorectal inflammation; Colorectal tumors
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and accounts for approximately 132,700 new cases and 49,700 deaths in the United States in 2015 [1]. Almost 55% of the CRC cases occur in more developed regions. The mortality of CRC is considerably lower (8.5% of the total) with more deaths (52%) in less developed regions of the world [2]. In the industrialized nations, the lifetime risk of developing CRCs is about 5%, and developing an adenoma, a non-cancerous colon tumor that can develop into CRCs, is 20% [3]. The human intestinal tract is colonized approximately 1014 CFU/g of microbiota. In recent years, it has been found that gut microbiota (including S. gallolyticus, E. Faecalis, Enterotoxigenic Bacteroides fragilis, F. nucleatum) is closely related to the occurrence of CRCs [3-7]. The colon mucosa is covered by a mucus layer that segregates the microbiota from the host colonic epithelium. Breaches of this protective mucus layer will lead to increased contact between mucosal microbiota and the colonic epithelial cells. Concomitant with increased access to the mucosal epithelium, microbial community communication, microbial metabolism, microbial structure and function are modified and often resulting in biofilm formation [8,9] (Figure 1). The direct contact of biofilms with epithelial cells results in perturbed epithelial metabolism and function, and facilitates chronic inflammation and even CRCs [8]. Compared with left-sided CRC, right-sided colon tumor has unique biological behavior (the “two-colon” concept) [10]. A recent study found that gut bacteria biofilm is widespread in the right-side colon tumors [11]. However, the role of biofilms in the carcinogenesis of CRC is still not clarified. The possible mechanism may include epithelial cell damage, DNA damage, chronic inflammation and bacteria carcinogens, etc.
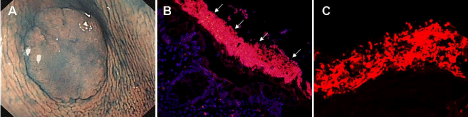
Figure 1: Detection of microbial biofilms on colon tumors.
A. Magnification chromoendoscopy showed the mucus layer on the tumor surface.
B. Fluorescence in situ hybridization (FISH) of all bacteria (red) on tumors. All were counterstained with the nuclear stain, DAPI (blue).
C. Confocal laser scanning microscope revealed the bacteria (red) in biofilms.
The structure of microbial biofilms
Biofilms form on abiotic or biotic surfaces and grow on a threedimensional structure. Biofilms are composed of host constituents, polysaccharides, cell-free enzymes, and bacteria embedded in a matrix of extracellular polymeric substances [12]. Many biofilms involve the production of an extracellular matrix (ECM), which encases the bacteria cells, binds the bacteria together, and can be composed of lipopolysaccharides, polysaccharides, proteins and extracellular DNA [13,14]. It is the ability of microorganisms to form biofilms that favor their persistence and survival.
The biofilm is more like a complex, highly differentiated, multicultural community much like our own city [15]. There are approximately 700 bacterial species in oral biofilm [16]. Decades ago, Listgarten and co-workers described the architecture of biofilms by light and electron microscopy on epoxy resin crowns and extracted teeth [17,18]. In a later study with hybridization (FISH), it was shown that subgingival biofilms formed on expanded polytetrafluoroethylene carriers which inserted into the depth of periodontal pockets [19]. In periodontitis, the subgingival biofilm is composed of Actinomyces sp., Tannerella forsythia, Fusobacterium nucleatum, Spirochaetes and Synergistetes. Streptococcus sp. and the yeast Candida albicans form structures in supragingival biofilm [20].The formation of a biofilm by H. pylori has been shown in acidic conditions of the stomach [21]. H. pylori strain TK1402, which is isolated from a patient with duodenal and gastric ulcers, has been shown to have strong biofilm forming ability both inside and outside the host [22-24]. The formation of a biofilm by Bacteroidetes and Firmicutes may be associated with adenomas and CRCs. Biofilms detected on surgically resected, normal tissues were composed of Bacteroidetes, Gammaproteobacteria and Lachnospiraceae. Biofilms identified on normal mucosa obtained at colonoscopy from patients without CRCs were similarly composed of Bacteroidetes and Lachnospiraceae. Colonic mucosal biopsies with or without biofilms from healthy individuals did not reveal any invasive bacteria [8]. F. nucleatum is a gram-negative oral symbiotic bacteria, which has the potential to be pathogenic, sometimes causes periodontal disease. In October 2011, two research teams from Canada BC Cancer Institute and Broad Institute confirmed that F. nucleatum was present in the gut, and its abundance was associated with CRCs [25,26]. Compared to normal tissue, FISH and qPCR analysis of Fusobacterium revealed a significant increase in CRCs tissues, and higher abundance in right side tumor location [8,27]. Recent research showed that F. nucleatum can modulate E-Cadherin/ β-Catenin signaling via its FadA adhesin to promote colorectal carcinogenesis [26].
The development of most of the biofilms occurs in five stages: (1) reversible aggregation of planktonic cells on a biotic or abiotic surface; (2) irreversible adhesion; (3) formation of microcolonies; (4) biofilm maturation; (5) detachment of cells and dispersion in a new niche [28]. Firstly, planktonic cells approach the surface and move slowly. The forces that mediate bacterial adhesion to surfaces, have been reasonably well identified in the past and include ubiquitously present attractive Lifshitz-van der Waals forces, acid-base bonding, electrostatic interactions, polysaccharides and several specific protein-protein interactions [28-30]. By providing more diverse adhesion sites for other microorganisms, the microbes that initially colonize the surface facilitate the development of the biofilm. As the microbial population of the incipient biofilm increases a polymeric matrix develops and microcolony forms [12]. Bacterial adhesion and aggregation are accelerated by force-generating organelles such as type IV pili and flagella, or some adhesion proteins [15,31,32]. Followed by bacterial division and production of the extracellular matrix and biofilm maturation, the ECM is disintegrated, and the bacteria detach the surface and disperse in a new niche [33]. Quorum sensing (QS) is a consistent communication mechanism between bacteria that influence gene expression. It is sensitive to changes in cell density. When the attached bacteria form microcolonies, the population density increases and quorum signals reach sufficient levels to activate the maturation and disassembly of the biofilm in a coordinated manner [33]. When nutrients and other resources become limited and waste products accumulate, biofilm dispersion is necessary to allow bacteria to escape and colonize new niches. There are different tactics to accomplish biofilm dispersion: ending the synthesis of the biofilm matrix compounds, degrading the matrix and disrupting noncovalent interactions between matrix components [34]. Biofilm formation is regulated by extracellular signaling (quorum sensing; homoserine lactone; cis-unsaturated fatty acid), and intracellular signaling (cyclic di-GMP; cyclic di-AMP; NO) [14].
Biofilms play important roles in human infections including inflammatory bowel disease (IBD), colorectal tumors, endocarditis, periodontitis, caries, otitis media, sinusitis, endophthalmitis, keratitis, chronic bacterial prostatitis, vaginitis, and lung infections in patients with cystic fibrosis [8,12,14,20,28,31,35-38]. Previous animal studies found that the biofilms mainly exist in the proximal colon [39,40]. Similarly, the biofilm on human appendix is the most significant. The biofilms gradually reduce from the appendix to the distal colon. Compared with an ascending and transverse colon, the biofilm on cecum is more obvious. However, no biofilms were detected on the left half colon [8,41]. In the different department, the thickness and density of biofilms are different. The intestinal bacterial biofilms were defined as massive bacterial invasions (>109 bacteria/mL) of the mucus layer spanning at least a linear distance of 200 μm from the epithelial surface [8]. The thickness of biofilm at the atmosphereliquid interface of in vitro cultivated microorgnanism collected from descending colon mucosa ranged from 20 to 80 μm. The anaerobic bacteria formed biofilms ranging from 2×103-2×105 cfu of culturable bacteria per peg [42]. Compared with paired normal colon tissues, biofilm depth was significantly increased in tumor samples, but the density did not differ between tumors (CRCs or adenomas) and their paired normal colon tissues [8]. These previous findings showed that biofilm mainly exists in the right colon, and the interaction between host epithelial cells and the microbes promotes its formation.
The effect of microbial biofilm on the intestinal epithelial cells
The mature of biofilm will change the epithelial cell biology, including the epithelial permeability, cell proliferation and apoptosis, gene transcription and protein expression [43-47].
Biofilm formation is associated with reduced or redistributed colonic epithelial cell E-cadherin, consistent with increased epithelial permeability, resulting increased contact between bacterial biofilm antigen and the colonic epithelial cells, perturbed epithelial function, and chronic inflammation and cancer [8]. Disruptions of biofilms and a concurrent increased release of pathogenic planktonic bacteria from these biofilms can damage tight junctions and increase epithelial permeability [48]. As the important components of EMC, the curli amyloid fibrils expressed by biofilm bacteria can activate the TLR2/PI3K pathway in intestinal epithelial cells, resulting in the reinforcement of the epithelial barrier and decreased epithelial permeability, preventing more bacteria to translocate to the basolateral side of the epithelium [49].
Biofilm can affect gene transcription and expression of the epithelial cells. By releasing antimicrobial peptides as well as cytokines in response to the diverse microorganisms establishing oral biofilms, oral epithelial cells change in transcript levels [46]. On the proteomic level, in vitro studies showed that more secreted proteins were downregulated, than upregulated after 24h, and this difference was further increased after 48h. The most significantly down-regulated proteins were actin, CD59 (human leukocyte antigenMIC11), annexin family proteins and cornulin. They are important in mucosal/epithelial immune response and epidermal differentiation [50]. Some proteins are closely related to the disassembly of the DNA complex, chromatin, and nucleosome, the disruption of the epithelial tissue integrity, and the inflammatory response. These changes may represent the aftermath of earlier-triggered apoptotic cascades [47,50]. However, there is little research about the effect of biofilm on gene expression of the intestinal epithelial cells. Studies have shown that in HBUS mice, the presence of biofilm bacteria can trigger gene expressions associated with a bactericidal activity such as Arg1, Ptgs2 (also known as COX2), Serpine1, Reg3b, and Reg3g [51]. A recent study reported that biofilm positive normal colon tissues in the patients with CRC displayed reduced E-cadherin expression in crypt epithelial cells, and increased IL-6 and STAT3 expressions [8].
In addition, the breakdown products of biofilm can also affect the epithelial cell metabolism. The bacteria in a biofilm can break down bile acid to form deoxycholic acid, and increased deoxycholic acid can activate NF - kB, enhance DNA synthesis, and result in epithelial cell proliferation and apoptosis [52].
Microbial biofilm and host inflammatory reactions
Biofilms have gotten increasing attention for illuminating dynamic and reciprocating interactions between the organisms in biofilms and human immune effector cells. It had been believed that immune responses were triggered primarily by antigens on the outer surface of the biofilm, with the matrix serving as a mechanical barrier to antibodies and immune cells and other proteins [53]. As the colon mucosa continued exposure to the intestinal bacterial biofilm and its metabolites, the immune response of epithelial cells would develop, including the production of Proinflammatory cytokines, chemokines, and the expression of matrix metalloproteinases. The interaction between host and biofilm cause an immune response. In periodontitis, plasma cells and lymphocytes are the predominant cells in the chronic inflammatory lesion, with the presence of B cells being proportionally larger than T cells. On the other hand, the immune response in IBD is mediated by T lymphocytes as a consequence of a genetic trait associated with T-cell deregulation [54].
Innate immune response: The innate immunity associated with biofilms includes Toll-like receptors (TLRs), antimicrobial peptides (AMPs), neutrophil, chemokines and cytokines. Within minutes or hours, the innate immunity is activated towards a variety of antigens and is mediated by several different effectors such as neutrophils, macrophages, monocytes, dendritic cells, NK cells and epithelial cells. It’s a widely non-specific reaction, primarily directed towards microbial antigens [54]. Host recognition of microorganism-associated molecular patterns (MAMPs) occurs via various pattern recognition receptors (PRRs; such as TLRs). MAMPs include lipopolysaccharide (LPS), flagellin and nucleic acids [55]. In particular, alterations in signaling of TLR4 have been linked to the progression of CRC [56]. The imbalance of biofilm structure can induce mild inflammation or the host systemic response and produce LPS, leading to metabolic endotoxemia.
Neutrophil Research has shown that biofilms are overlaid or surrounded with neutrophils but not penetrated and actively killed by the neutrophils [57]. One explanation may be that these neutrophils are constantly being recruited by the biofilm [57]. Through their rhamnolipid protective shield, bacteria in biofilms may protect themselves from being phagocytized by neutrophils [57]. However, other biofilm matrix components, including bacterial DNA and alginate, were reported to stimulate neutrophils [57,58]. Bacteria resident in biofilms evidence a detachment response that releases a cloud of bacteria from the biofilm to envelop attracted and homing neutrophils and obscure their targets, so phagocytosis alone is ineffective against biofilm residing bacteria [53]. A recent study found that neutrophils were closely related to the occurrence of serrated polyps [51].
The cytokines and growth factors are often associated with the presence of biofilm bacteria and their endotoxins. In chronic wounds, the chronic S. aureus biofilm infection indicates a predominantly Th1 and Th17 type response, and increased levels of IL-2, IL-4, IL-6, IL-12, IL-17, and TNF-a [57]. Th17 cells can secrete Proinflammatory factor IL-17 and IL-22, resulting in tissue damage [59]. In human periodontitis lesions, there are increasing evidence of the presence of Th17 and IL-17 cells, which may be associated with disease severity [60]. In the gut mucosa of IBD patients, the role of innate immunity appears to be crucial, because defects of the innate immune response trigger changes in the Th1 and Th2 cell responses. These disruptions of the regulatory T cells contribute to altering the mucosal immunity [54]. Respectively, Crohn’s disease(CD) demonstrates a characteristic Th1 type of immune response, dominated by overproduction of IFN-g, while ulcerative colitis (UC) is characterized by an atypical Th2 response, with elevated production of IL-13 [61,62]. In the serrated polyps of HBUS mice, biofilm bacteria invade into the lamina propria. Compared with unaffected cecal tissue, the Proinflammatory cytokines IL1-α, IL1-β and Tnf, and the chemokines Ccl1, Ccl2, Ccl17, Cxcl2 and Cxcl16 were significantly upregulated [51] (Figure 2).
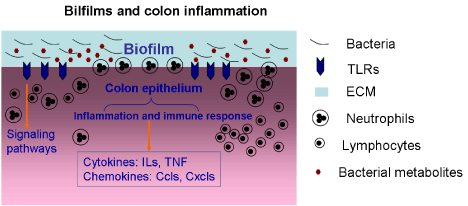
Figure 2: Microbial biofilms and colonic inflammatory reactions. TLRs, Tolllike
receptors. ECM, extracellular matrix.
Microbial biofilms in colorectal carcinogenesis
Biofilm has been implicated in nonmalignant diseases such as inflammatory bowel disease, and recent evidence suggests that the biofilms in the colon were associated with increased risk of CRC [8,55,63]. Complex bacterial communities invade to colonize the mucus layer of the colonic mucosa and encased in mucus, which was identified in nearly all colorectal tumors (cancers and adenomas), especially proximal CRCs [8].
However, the precise mechanisms by which this process happens are still not fully elucidated. The gut microbiota potentially contributes to host cancer risk via three major routes: (1) altering host cell proliferation or turnover; (2) influencing immune function; (3) metabolizing ingested and host-derived products [3].
Colonic biofilms alter the cancer metabolome to produce a regulator of cellular proliferation, and potentially affect cancer development and progression. Increased polyamine concentrations are associated with eukaryotic proliferation, and polyamines are oncometabolites that regulate the LIN28/let-7 pathway in CRCs [64,65]. In colon cancer tissues positive for the presence of biofilms, an overproduction of N1, N12-diacetylspermine and acetylated polyamine was observed [63]. Other metabolites that were significantly changed included dodecanoic acid, undecanoic acid, isobutrylcarnitine, capryloylglycine, and ornithine decarboxylase in human cancer cell lines [63,66].
Gut bacteria biofilms drive inflammation within the colon, and such inflammation is strongly linked to CRCs. Enhanced epithelial permeability facilitates Proinflammatory cytokine production and bacterial antigen translocation [67]. In biofilm-positive compared to biofilm-negative normal tissues from the cancer host, an increase in both IL-6 and STAT3 activation was seen [8].
No single bacterial species has been identified as a risk factor for CRC, but recent studies reported an increase in the abundance of Fusobacterium nucleatum (Fn) in human colorectal tumors compared to controls [4,5,25,27]. However, these studies only show that Fn is related with the incidence of colorectal tumors, but do not reveal that it is the cause or the result of CRC. Fn was detected in 24% of hyperplastic polyps, 30% of traditional serrated adenomas, 35% of sessile serrated adenomas, 33% of non-serrated adenomas, and 56% of CRCs [68]. Fn may contribute to the progression of CRC, and is associated with microsatellite instability (MSI) and CpG island methylator phenotype (CIMP) [6,68]. Fn was localized in the mucus layer of the epithelium as well as within the colonic crypts [27]. Through its unique FadA adhesin, Fn adheres to, invades, and induces inflammatory and oncogenic responses to stimulate the growth of CRC cells [26]. FadA binds to E-cadherin activates β-catenin signaling and regulates the inflammatory and oncogenic responses [26]. In addition, Bacteroides fragilis, S. gallolyticus, E. coli NC101 and Methanobacteriales are also associated with the occurrence of CRC [52,69-72].
It’s found that higher levels of Fn were related to lower levels of Faecalibacterium prausnitzii [71]. As the butyrate-producing bacteria and a healthy biomarker in the prevention of CRCs, F. prausnitzii levels decreased significantly in CRC patients compared to healthy subjects [73]. Another healthy biomarker is Bifidobacterium. It has been reported that lower levels of Bifidobacterium spp. in fecal have been related to CRCs compared to healthy controls [74].
The management of microbial biofilms
The formation of microbial biofilms is an important reason for the failure of antimicrobial therapy. For example, Vancomycin is the most commonly drug for S. aureus biofilm infections. However, the administration of this drug owns to the propensity of S. aureus to develop resistance [75]. Compared to the minimum inhibitory concentration against planktonic bacterial cells, effective antibiotic concentrations against biofilm may be many times higher, and the bacterial counts are generally only temporarily suppressed [57]. Several mechanisms play an important role in biofilm survival, including protection provided by matrix polysaccharides, biofilmspecific protection against oxidative stress and biofilm-specific expression of efflux pumps [28,76]. ECM that encases bacteria can increase bacterial stability and survival, protecting the bacteria from the action of antimicrobial agents, host immune responses, bacteriophages and phagocytic amoeba [28,77].
Studies found that the production of reactive oxygen species (ROS) is involved in the killing of bacteria under certain conditions [78]. Protection against oxidative stress decreases the activity of bactericidal drugs, and this protection is important for the survival of treated biofilms. Besides, the matrix polysaccharides limit the penetration of antibiotics into biofilms [79]. Electrostatic repulsion as well as the hydrophobic help to prevent polarly and charged antibiotics from reaching the inner regions of a biofilm community. Daptomycin, a cyclic lipopeptide molecule, disrupts the cytoplasmic membrane of bacteria, resulting in rapid depolarization and cessation of DNA and RNA synthesis. It has been shown to be able to penetrate S. epidermidis biofilm rapidly [75,80]. In addition, bacteria resistant to antimicrobial agents can transfer the genes for resistance to neighboring receptive bacteria. Such promiscuous gene transfer can convert a previously avirulent commensal organism into a highly virulent pathogen [53,81]. Besides, the development of subpopulations of dormant and metabolically less active ‘‘persister’’ bacteria can reduce the efficacy of bacteriostatic antimicrobial agents [82,83].
The frequent failure of antibiotic therapy led researchers to look for alternative methods with a mechanism of action different from that of antibiotics. At present, quorum sensing is a new target for the development of antibacterial agents [84]. Both in vitro and in vivo, quorum-sensing inhibitors increase the susceptibility of bacterial biofilms to existing antibiotics [85]. Antimicrobial peptides (AMPs) as a potential treatment, is recently attracting more attention. AMPs result in a low rate of induced resistance, crucial against biofilms, and in efficacy against a wide range of microorganisms and is particularly suitable to treat biofilms with the polymicrobial character [86].
Other mechanisms of AMPs include binding with DNA, inhibition of protein synthesis, detoxification of LPS, and interaction with polysaccharide components of the matrix and disaggregate biofilms [87-89]. Moreover, bacteriophages may both prevent biofilm formation and contribute to the eradication of biofilm bacteria by facilitating the degradation of extracellular polymeric substances (EPS), the permeation of bacteriophages into deeper biofilm layers and lysis of the susceptible bacterial cells [90].
It’s conceivable that patients with the colorectal inflammatory disease or CRC risk might benefit from eradicating microbial biofilms because biofilms are closely related to colorectal inflammation and carcinogenesis. However, few researchers have studied the management of colorectal biofilms. Further studies are needed to investigate the therapy of colorectal biofilms and following benefits in patients with the colorectal inflammatory disease or CRC risk in the future.
In conclusion, microbiota biofilm is a feature in colorectal inflammation and CRCs, which is mainly composed of gut microbiota, host constituents, polysaccharides, cell-free enzymes and ECM. Biofilm interacts with the gut epithelium and immune system, leading to inflammation and carcinogenesis of the colorectum. Progress in the management of microbial biofilms has been achieved in recent years.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015; 65: 5-29.
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015; 136: E359-386.
- Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014; 15: 317-328.
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012; 22: 292-298.
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012; 22: 299-306.
- Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014; 74: 1311-1318.
- Jobin C. Colorectal cancer: looking for answers in the microbiota. Cancer Discov. 2013; 3: 384-387.
- Dejea CM, Wick EC, Hechenbleikner EM, White JR, Mark Welch JL, Rossetti BJ, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci U S A. 2014; 111: 18321-18326.
- Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011; 1: 4659-4665.
- Papagiorgis P. Colorectal cancer: dichotomous or continuum model? Perhaps, a combination of both. Gut. 2013; 62: 1519-1520.
- Bauer KM, Hummon AB, Buechler S. Right-side and left-side colon cancer follow different pathways to relapse. Mol Carcinog. 2012; 51: 411-421.
- Torresyap V, Moshaverinia A, Chee WW. Biofilms in restorative dentistry: A clinical report. J Prosthet Dent. 2015; 113: 524-527.
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010; 8: 623-633.
- Römling U, Kjelleberg S, Normark S, Nyman L, Uhlin BE, Åkerlund B. Microbial biofilm formation: a need to act. J Intern Med. 2014; 276: 98-110.
- Watnick P, Kolter R. Biofilm, city of microbes. J Bacteriol. 2000; 182: 2675-2679.
- Wang W, Tao R, Tong Z, Ding Y, Kuang R, Zhai S, et al. Effect of a novel antimicrobial peptide chrysophsin-1 on oral pathogens and Streptococcus mutans biofilms. Peptides. 2012; 33: 212-219.
- Listgarten MA, Mayo HE, Tremblay R. Development of dental plaque on epoxy resin crowns in man. A light and electron microscopic study. J periodontol. 1975; 46: 10-26.
- Listgarten MA. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J Periodontol. 1976; 47: 1-18.
- Wecke J, Kersten T, Madela K, Moter A, Göbel UB, Friedmann A, et al. A novel technique for monitoring the development of bacterial biofilms in human periodontal pockets. FEMS Microbiol Lett. 2000; 191: 95-101.
- Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmür R, et al. Oral biofilm architecture on natural teeth. PLoS One. 2010; 5: e9321.
- Cammarota G, Branca G, Ardito F, Sanguinetti M, Ianiro G, Cianci R, et al. Biofilm demolition and antibiotic treatment to eradicate resistant Helicobacter pylori: a clinical trial. Clin Gastroenterol Hepatol. 2010; 8: 817-820 e813.
- Carron MA, Tran VR, Sugawa C, Coticchia JM. Identification of Helicobacter pylori biofilms in human gastric mucosa. J Gastroint Surg. 2006; 10: 712-717.
- Yonezawa H, Osaki T, Kurata S, Fukuda M, Kawakami H, Ochiai K, et al. Outer membrane vesicles of Helicobacter pylori TK1402 are involved in biofilm formation. BMC Microbiol. 2009; 9: 197.
- Yonezawa H, Osaki T, Woo T, Kurata S, Zaman C, Hojo F, et al. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe. 2011; 17: 388-390.
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013; 14: 207-215.
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013; 14: 195-206.
- McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013; 8: e53653.
- Bispo PJ, Haas W, Gilmore MS. Biofilms in infections of the eye. Pathogens. 2015; 4: 111-136.
- Busscher HJ, Rinastiti M, Siswomihardjo W, van der Mei HC. Biofilm formation on dental restorative and implant materials. J Dent Res. 2010; 89: 657-665.
- Beaussart A, Herman P, El-Kirat-Chatel S, Lipke PN, Kucharíková S, Van Dijck P, et al. Single-cell force spectroscopy of the medically important Staphylococcus epidermidis-Candida albicans interaction. Nanoscale. 2013; 5: 10894-10900.
- Lee JH, Regmi SC, Kim JA, Cho MH, Yun H, Lee CS, et al. Apple flavonoid phloretin inhibits Escherichia coli O157:H7 biofilm formation and ameliorates colon inflammation in rats. Infect Immun. 2011; 79: 4819-4827.
- Krzysciak W, Jurczak A, Koscielniak D, Bystrowska B, Skalniak A. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014; 33: 499-515.
- Solano C, Echeverz M, Lasa I. Biofilm dispersion and quorum sensing. Curr Opin Microbiol. 2014; 18: 96-104.
- Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013; 64: 175-188.
- Hamilos DL. Host-microbial interactions in patients with chronic rhinosinusitis. J Allergy Clin Immunol. 2014; 133: 640-653.
- Russell PT, Bekeny JR. Oral antibiotics and the management of chronic sinusitis: what do we know?. Curr Opin Otolaryngol Head Neck Surg. 2014; 22: 22-26.
- Coticchia JM, Chen M, Sachdeva L, Mutchnick S. New paradigms in the pathogenesis of otitis media in children. Front Pediatr. 2013; 1: 52.
- Schilling J, Loening-Baucke V, Dörffel Y. Increased Gardnerella vaginalis urogenital biofilm in inflammatory bowel disease. J Crohns Colitis. 2014; 8: 543-549.
- Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005; 11: 1131-1140.
- Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR. Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol. 2004; 28: 23-27.
- Randal Bollinger R, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J Theor Biol. 2007; 249: 826-831.
- Sproule-Willoughby KM, Stanton MM, Rioux KP, McKay DM, Buret AG, Ceri H. In vitro anaerobic biofilms of human colonic microbiota. J Microbiol Methods. 2010; 83: 296-301.
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012; 491: 254-258.
- Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009; 15: 79-80.
- Wang L, Yi T, Kortylewski M, Pardoll DM, Zeng D, Yu H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009; 206: 1457-1464.
- Langfeldt D, Neulinger SC, Stiesch M, Stumpp N, Bang C, Schmitz RA, et al. Health- and disease-associated species clusters in complex natural biofilms determine the innate immune response in oral epithelial cells during biofilm maturation. FEMS Microbiol Lett. 2014; 360: 137-143.
- Thurnheer T, Belibasakis GN2, Bostanci N3. Colonisation of gingival epithelia by subgingival biofilms in vitro: role of "red complex" bacteria. Arch Oral Biol. 2014; 59: 977-986.
- Motta JP, Flannigan KL, Agbor TA, Beatty JK, Blackler RW, Workentine ML, et al. Hydrogen sulfide protects from colitis and restores intestinal microbiota biofilm and mucus production. Inflamm Bowel Dis. 2015; 21: 1006-1017.
- Oppong GO, Rapsinski GJ, Newman TN, Nishimori JH, Biesecker SG, Tükel Ç. Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect Immun. 2013; 81: 478-486.
- Bostanci N, Bao K, Wahlander A, Grossmann J, Thurnheer T, Belibasakis GN. Secretome of gingival epithelium in response to subgingival biofilms. Mol Oral Microbiol. 2015; 30: 323-335.
- Bongers G, Pacer ME, Geraldino TH, Chen L, He Z, Hashimoto D, et al. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J Exp Med. 2014; 211: 457-472.
- Abdulamir AS, Hafidh RR, Bakar FA. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer. 2010; 9: 249.
- Hall MR, McGillicuddy E, Kaplan LJ. Biofilm: basic principles, pathophysiology, and implications for clinicians. Surg Infect (Larchmt). 2014; 15: 1-7.
- Indriolo A, Greco S, Ravelli P, Fagiuoli S. What can we learn about biofilm/host interactions from the study of inflammatory bowel disease. J Clin Periodontol. 2011; 38 Suppl 11: 36-43.
- Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014; 12: 661-672.
- Neal MD, Sodhi CP, Jia H, Dyer M, Egan CE, Yazji I, et al. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem. 2012; 287: 37296-37308.
- Zhao G, Usui ML, Lippman SI, James GA, Stewart PS, Fleckman P, et al. Biofilms and Inflammation in Chronic Wounds. Adv Wound Care (New Rochelle). 2013; 2: 389-399.
- Seth AK, Geringer MR, Gurjala AN, Abercrombie JA, Chen P, You T, et al. Understanding the host inflammatory response to wound infection: an in vivo study of Klebsiella pneumoniae in a rabbit ear wound model. Wound Repair and Regen. 2012; 20: 214-225.
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007; 25: 821-852.
- Cheng WC, Hughes FJ, Taams LS. The presence, function and regulation of IL-17 and Th17 cells in periodontitis. J Clin Periodontol. 2014; 41: 541-549.
- Cobrin GM, Abreu MT. Defects in mucosal immunity leading to Crohn's disease. Immunol Rev. 2005; 206: 277-295.
- Targan SR, Karp LC. Defects in mucosal immunity leading to ulcerative colitis. Immunol Rev. 2005; 206: 296-305.
- Johnson CH, Dejea CM, Edler D, Hoang LT, Santidrian AF4, Felding BH4, et al. Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 2015; 21: 891-897.
- Gerner EW, Meyskens FL Jr. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004; 4: 781-792.
- Paz EA, LaFleur B, Gerner EW. Polyamines are oncometabolites that regulate the LIN28/let-7 pathway in colorectal cancer cells. Mol Carcinog. 2014; 53 Suppl 1: E96-106.
- Linsalata M, Cavallini A, Messa C, Orlando A, Refolo MG, Russo F. Lactobacillus rhamnosus GG influences polyamine metabolism in HGC-27 gastric cancer cell line: a strategy toward nutritional approach to chemoprevention of gastric cance. Curr Pharm Des. 2010; 16: 847-853.
- Hold GL, Garrett WS. Gut microbiota. Microbiota organization--a key to understanding CRC development. Nat Rev Gastroenterol Hepatol. 2015; 12: 128-129.
- Ito M, Kanno S, Nosho K, Sukawa Y, Mitsuhashi K1, Kurihara H1, et al. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015; 137: 1258-1268.
- Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009; 22: 349-369, Table of Contents.
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012; 338: 120-123.
- Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015; 50: 167-179.
- Holma R, Osterlund P, Sairanen U, Blom M, Rautio M, Korpela R. Colonic methanogenesis in vivo and in vitro and fecal pH after resection of colorectal cancer and in healthy intact colon. Int J Colorectal Dis. 2012; 27: 171-178.
- Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013; 66: 462-470.
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008; 57: 1470-1481.
- Bhattacharya M, Wozniak DJ, Stoodley P. Prevention and treatment of Staphylococcus aureus biofilms. Expert Rev Anti Infect Ther. 2015; 13: 1499-1516.
- Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014; 22: 326-333.
- Percival SL, Suleman L. Biofilms and Helicobacter pylori: Dissemination and persistence within the environment and host. World J Gastrointest Pathophysiol. 2014; 5: 122-132.
- Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013; 339: 1213-1216.
- Lewis K. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol. 2008; 322: 107-131.
- Stewart PS, Davison WM, Steenbergen JN. Daptomycin rapidly penetrates a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 2009; 53: 3505-3507.
- Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol. 2012; 65: 183-195.
- Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob Agents Chemother. 2004; 48: 2659-2664.
- Lewis K. Persister cells: molecular mechanisms related to antibiotic tolerance. Handb Exp Pharmacol. 2012; 121-133.
- Li YH, Tian X. Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel). 2012; 12: 2519-2538.
- Brackman G, Cos P, Maes L, Nelis HJ, Coenye T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother. 2011; 55: 2655-2661.
- Batoni G, Maisetta G, Brancatisano FL, Esin S, Campa M. Use of antimicrobial peptides against microbial biofilms: advantages and limits. Curr Med Chem. 2011; 18: 256-279.
- Zhang L, Hinz AJ, Nadeau JP, Mah TF. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J Bacteriol. 2011; 193: 5510-5513.
- Park SC, Park Y, Hahm KS. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int J Mol Sci. 2011; 12: 5971-5992.
- Di Luca M, Maccari G, Nifosì R. Treatment of microbial biofilms in the post-antibiotic era: prophylactic and therapeutic use of antimicrobial peptides and their design by bioinformatics tools. Pathog Dis. 2014; 70: 257-270.
- Parasion S, Kwiatek M, Gryko R, Mizak L, Malm A. Bacteriophages as an alternative strategy for fighting biofilm development. Pol J Microbiol. 2014; 63: 137-145.