
Research Article
Austin J Genet Genomic Res. 2015;2(1): 1008.
Validation of HMG CoA Reductase as Internal Control for Hazelnut Pollen Allergens Expression Analysis
Jana Žiarovská1*, Natalia Nikolaieva2, Katerina Garkava² and Ján Brindza¹
¹Department of Genetics and Plant Breeding, Slovak University of Agriculture in Nitra, Slovak Republic
²Institute of Ecological Safety, National Aviation University, Ukraine
*Corresponding author: Žiarovská J, Department of Genetics and Plant Breeding, Faculty of Agrobiology and Food Resources, Slovak University of Agriculture in Nitra, Tr. A. Hlinku 2, 94976, Slovak Republic
Received: August 19, 2014; Accepted: February 10, 2015; Published: February 13, 2015
Abstract
In the study, 3-hydroxy-3-methylglutaryl coenzyme A reductase gene was validated for its application in hazelnut pollen allergens CorA and profillin expression analysis. HMG CoA reductase was used as internal control in expression analysis by one-step real-time PCR approach. Pollen sample from rural area from Sumy region (Ukraine) was used as the calibrator and the sample from Kamenets-Podolskiy (cement plant area; Ukraine) was chosen for the expession analysis. Using one-step real time PCR approach, following values of expression level were obtained in this study: 0,63 x higher for CorA and 51,6 higher x for profilin hazelnut pollen ellergen. The results and parameters of expression profile data are comparable to the two-step real-time PCR, that were previously reported for Corylus avelana, L. pollen allergens and confirmed HMG CoA reductase as suitable internal control for the CorA and profillin expression analysis.
Keywords: Corylus avelana, L; HMG CoA reductase; Pollen allergens; Expression; RT-PCR
Abbreviations
HMG CoA Reductase: 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase; mRNA: messenger Ribonucleic Acid; NCBI: National Centre of Biotechnology Information; RT-PCR: Reverse Transcribed Polymerase Chain Reaction; RT-qPCR: Real-Time quantitative Polymerase Chain Reaction
Introduction
Respiratory diseases with a background in pollen allergens possess an increased tendency throughout the urbanized area. Pollen allergic diseases are the result of hypersensitivity to aeroallergens and are reported as to affecting up to the 30% of population [1,2]. Corylus avellana is a known source of food allergy, but the immunological cross-reactivity of hazel pollen is reported and known as well [3-5]. Two pollen allergens are described for Corylus avellana – CorA and pollen profilin [6]. Both of them were reported as to be related to the birch pollen allergens BetV1 and BetV2 [7]. For pollen allergens, a discrepancy between the number of pollen grains and quantities of pollen allergen in the atmosphere was reported by Takahashi et al. and Rantio-Lehtimaki et al. [8,9]. Thinking of above mentioned circumstances, developing of reproducible and reliable methods of the allergenic potencial of different sources of spring pollinosis will lead to their better management.
Gene expression analysis are nowadays an inevitable part of routine analysis in many fields. The quantitation of mRNA transcripts of expressed genes is performed in real-time quantitative PCR (RTqPCR). Data obtained in the quantitation, after its normalization, provide a powerful source of gene expresion patterns [10]. Different housekeeping genes are used as internal controls for RT-qPCR analysis [11-13]. Comparison of hazelnut pollen expression level using 18S rRNA as internal control in quantitative real-time PCR was performed previously by Ražná et al. [14]. Using more than one internal control is suggested in RTqPCR to get more reliable results [15]. Here, the 3-hydroxy-3-methylglutaryl coenzyme A reductase was validated for hazelnut pollen allergens expression analysis.
In plant, 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase (HMG CoA reductase) is involved in catalysis of isoprenoid biosynthesis. It catalyses the synthesis of mevalonate, that belongs to molecules taking part in growth regulators, photosynthetic pigments, mitochondrial electron transfer chain components, dolichon, phytoalexin and natural rubber [16]. Plant HMG CoA reductase has a vital role in the mevalonate pathway, critical in regulation of normal plant development and adaptation to demanding environmental conditions [17]. As reported by Leivar et al. [17], protein isoforms of HMG CoA reductase gene posses different roles in plant tissues. The HMGR1S transcripts are found elsewhere in plants and in high levels during the first stages of development and in inflorescences [18]. HMGR1L and HMGR2 transcripts are detected only in selected tissues and are much more less abundant than the HMGR1S mRNA [19,20]. These suggest a housekeeping role for HMGR1S and a specialized role for HMGR1L and HMGR2 [17].
The aim of the study was to validate 3-hydroxy-3-methylglutaryl coenzyme A reductase gene for its application in allergens expression analysis of hazelnut pollen allergens CorA and profillin. HMG CoA reductase was used as internal control in expression analysis by onestep real-time PCR approach.
Materials and Methods
Pollen material and treatment
Pollen sample was obtained in two different places in Ukraine (Figure 1). The sample from village area from Sumy region was used as the calibrator and the sample from Kamenets-Podolskiy (cement plant area) was chosen for the expession analysis. Pollen was obtained after anther dehiscence. The samples were immediately desicated and stored in aseptic container until allergen expression analysis.
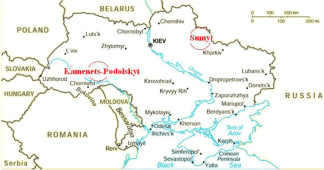
Figure 1: Localization of pollen samples analysed in the study.
Sample preparation protocol
One step real-time PCR approach was used for analysis. Total RNA was extracted from pollen by GeneLET™ Plant RNA Purification Mini Kit (Thermo Scientific) following manufacturers instructions. Quality and quantity of extracted RNA was checked using NanoPhotometer® (Implen) and samples were diluted to 20 ng of total RNA for further analysis. The iScript one-step RT-PCR kit with SYBR® Green (Bio-Rad Laboratories) was used to perform analysis in total volume of 25 μl.
Expression analysis
The mRNA sequences of studied genes were obtained from NCBI database. Bioinformatic evaluation of nucleotide data was applied for HMG CoA reductase. Primers for HMG CoA reductase (accession in NCBI - KF306244 and EF206343) were designed using Primer-Blast software under NCBI database. The sequences were as follows: forward primer 5´ gtcctcaaaaccaacgtggc 3´; reverse primer 5´acattctgagcggggtcttg 3´. Primers for CorA and profilin were designed as reported previously [14]. PCR was performed in CFX96 cycler (Bio-Rad Laboratories). Real-time PCR reaction for HMG CoA reductase expression analysis was optimized and validated for previously reported protocol of hazelnut pollen allergens [14]. Primer and RNA concentrations were optimized for their compactibility with the temperature and time conditions reported for CorA and profilin expression analysis. Primer dimers formation was evaluated by melting curve analysis. The CFX96 software generated standard curve by plotting Ct versus concentrations logarithms for each control. Hazelnut pollen allergens quantitations were processed using Pfaffl method [21]. Allergens expession was normalized agains 3-hydroxy-3-methylglutaryl coenzyme A reductase gene used as internal control. Validating of HMG CoA reductase results were performed by comparing them to the results of our previous study [14], where two-step real-time PCR approach was used for expression analysis of the same samples and 18S rRNA as internal control.
Results and Discussion
In the present study, using of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene as internal control were performed for utilizing it in hazelnut pollen allergens analysis. The one-step realtime PCR approach was chosen to simplifying the process of analysis. Obtaining efficient protocol without timeconsuming analysis is important for all inhalant allergies sources not only from medicinal, but from their management point of view, too. Pollen allergens still remain one of the major sources of respiratory diseases [22,23], but the protocols for their expression level analysis are reported in literature very rare [14,24].
HMGR expression analysis optimization
Using of 18S rRNA as internal control was reported previously for hazelnut pollen allergens expression analysis by Ražná et al. [14]. In the NCBI database, two sequence of HMG CoA reductase are available. Observations about HMG CoA reductase suggest a housekeeping role for HGR1S as refereed in Leivar et al. [17]. Fist, in silico analysis were performed to compare both of sequences available in public database. Performing a BLAST analysis [25] agains the individual sequences of both accessions, the region with a full match were identified and this region was used for primer designing (Table 1).
primer sequence
matching sequence for EF206343 in NCBI
matching sequence for KF306244 in NCBI
5´gtcctcaaaaccaacgtggc3´
1345 - 1364
1213 – 1232
5´acattctgagcggggtcttg 3´
1483 - 1502
1351 - 1370
Table 1: Matching of primers used in the study with HMG CoA reductase sequences in public database.
Optimization of RNA content, primer content and annealing temperature in HMG CoA reductase expression analysis was performed. Using a temperature gradient 54 °C – 64 °C, the 58 °C (Figure 2) was showed to be an optimal for HMG CoA reductase amplicons generating. Tested concentration range for RNA content (5,10,20 and 40 ng) and primer concentration (200, 300, 400, 500 nmol. dm-3 and their combinations in forward and reverse position) showed the optimal for 20 ng of RNA content and 400 nmol. dm-3 for forward/reverse primer concentration.
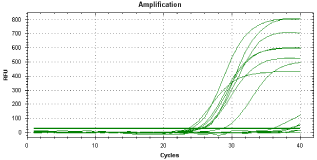
Figure 2: Effect of different annealing temperatures in HMG CoA reductase
amplification.
Standart curve and relative expression of hazelnuts pollen allergens using one-step real-time PCR approach
Five serial dillutions of RNA were used to obtain standart curves. Achieving efficiency was 98.9% for HMG CoA reductase, 99% for CorA allergen and 96% for profilin allergen. All constructed standart curves showed a linear regression between input RNA content and Ct values in three independent assays. The specifity of amplicons were inspected using melt analysis, where obtained values were 83.4;°C for CorA allergen and 79°C for profilin allergen (Figure 3). In this study the expression level of CorA and profillin hazelnut pollen allergens were prooved for a sample from a urbanized area of Kamenets- Podolskiy, cement plant area. The major allergens of hazel pollen were identified as to be molecular related to the birch pollen allergen BetV1 and birch profilin BetV2 [6,26,27].
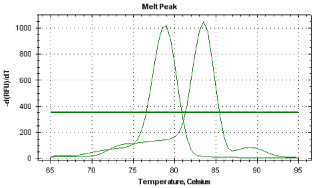
Figure 3: Melting point analyse of CorA and profilin amplicons.
Expression of allergens from tested sample were compared to the calibrator from village Sumy region. This combination was chosen, because the chemotactic activity and expression of BetV1 type allergen was reported to be higher in pollen samples from urban areas when comparing them to those from rural sites [24,28].
Previously, changes of CorA and profilin expression were reported as to be 0.62 and 52.9 x higher in the pollen sample from Kamenets- Podolskiy when compared them to the sample from rural area [14]. Using a one-step real time PCR approach, very similar values of expression level were obtained in this study: 0.63 x higher for CorA and 51.6 higher x for profilin hazelnut pollen ellergen. In spite of very similar expression level achieved by both used techniques, one-step and two-step relative quantitation, different Ct values were in analysis for CorA allergen (Table 2), as a result of differences among the types of nucleic acids used in analysis. For profilin, obtained data were again very similar.
Pollen allergen/
RT-PCR approachCorA
profilin
Ct1*
Ct2*
Ct3*
Ct1*
Ct2*
Ct3*
one-step real time
16,44
16,53
16,48
26,53
26,86
26,45
two-step real time+
20,04
19,4
19,57
26,93
25,95
26,7
*samples in triplicate; + data as obtained in authors´ previous study [14]
Table 2: Obtained Ct values in one-step and two-step real-time PCR based analysis of hazelnuts pollen allergens expression for the sample from urbanized area.
In plants, different genes used as internal control in real-time expression analyses have been identified [29,30]. The reliability of comparisons among expression levels of some samples´ genes depends on the normalization through internal controls, that are selected from housekeeping genes that shoul express in a stable way in selected plant tissues and given experimental conditions [10,31]. For their uniform expression, ubiquitin, 18S rRNA, β-tubulin, actin or glyceraldehyde-3-phosphate dehydrogenase are used widely in normalization of expression data [32,33]. The choise of housekeeping gene for using it as internal control leads to the accurate quantitative data analysis [10,30,34]. Here, the expression data were obtained and compared to those that were analysed using 18S rRNA for data normalization. 18S rRNA is used widely for this purpose [13,35] and was confirmed as reliable for different cell systems [10,11]. The results using both of them, 18S rRNA and HMG CoA reductase, as internal control showed to be consistent and reliable for quantitation of hazelnut pollen allergens expression levels.
Conclusion
Pollen-induced respiratory allergies became a part of many peoples lives in urbanized areas throughout the world. Here, the HMG CoA reductase is reported as suitable internal control for hazelnut pollen allergens expression level analysis. It has not beed reported as internal control in expression analysis yet. The potential use of HMG CoA reductase as internal control was analysed in expression profiles of pollen samples from Ukraine. Our findings confirm its potential to do so in hazelnut pollen allergen expression.
Acknowledgement
The research was supported by the project KEGA 001SPU/4-2012 Plant Genetic Technologies.
References
- Nielsen NH, Svendsen UG, Madsen F, Dirksen A. Allergen skin test reactivity in an unselected Danish population. The Glostrup Allergy Study, Denmark. Allergy. 1994; 49: 86-91.
- Midoro-Horiuti T, Goldblum RM, Kurosky A, Wood TG, Brooks EG. Variable Expression of Pathogenesis-Related Protein Allergen in Mountain Cedar (Juniperus ashei) Pollen. J Immunol. 2000; 164: 2188–2192.
- Rohac M, Birkner T, Reimitzer I, Bohle B, Steiner R, Breitenbach M, et al. The immunological relationship of epitopes on major tree pollen allergens. Mol Immunol. 1991; 28: 897–906.
- Ipsen H, Wihl JA, Petersen BN, Lowenstein H. Specificity mapping of patients IgE response towards the tree pollen major allergens Aln g I, Bet v I and Cor a I. Clin Exp Allergy. 1992; 22: 391–399.
- Heiss S, Fischer S, Müller WD, Weber B, Hirschwehr R, Spitzauer S, et al. Identification of a 60 kD cross-reactive allergen in pollen and plant derived food. J Allergy Clin Immunol. 1996; 98: 938–947.
- Hirschwehr R, Valenta R, Ebner C, Ferreira F, Sperr WR, Valent P, et al. Identification of common allergenic structures in hazel pollen and hazelnuts: a possible explanation for sensitivity to hazelnuts in patients allergic to tree pollen. J Allergy Clin Immunol. 1992; 90: 927–936.
- Lüttkopf D, Müller U, Skov PS, Ballmer-Weber BK, Wüthrich B, Skamstrup Hansen K, et al. Comparison of four variants of a major allergen in hazelnut (Corylus avellana) Cor a 1.04 with the major hazel pollen allergen Cor a 1.01. Molecular Immunology. 2002; 38: 515–525.
- Takahashi Y, Sakaguchi M, Inouye S, Miyazawa H, Imaoka K, Katagiri S. Existence of exine-free airborne allergen particles of Japanese cedar (Cryptomeria japonica) pollen. Allergy. 1991; 46: 588–593.
- Rantio-Lehtimaki A, Viander M, Koivikko A. Airborne birch pollen antigens in different particle sizes. Clin Exp Allergy. 1994; 24: 23–28.
- Aman S, Haq NU, Shakeel SN. Identification and validation of stable internal control for heat induced gene expression of Agave Americana. Pak J Bot. 2012; 44: 1289-1296.
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005; 56: 2907-2914.
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Bioph Res Comm. 2006; 345: 646-651.
- Garg R, Sahoo A, Tyagi AK, Jain M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Bioch Bioph Res Comm. 2010; 396: 283-288.
- Ražná K, Bežo M, Nikolaieva N, Garkava K, Brindza J, Žiarovská J. Variability of Corylus avellana, L. CorA and profilin pollen allergens expression. J Envir Sci Health B. 2014; 49: 639-645.
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002; 3.
- Chye ML, Tan CT, Chua NH. Three genes encode 3-hydroxy-3-methylglutaryl-coenzyme A reductase in Hevea brasiliensis: hmg1 and hmg3 are differentially expressed. Plant Mol Biol. 1992; 19: 473-484.
- Leivar P, Antoli´n-Llovera M, Ferrero S, Closa M, Arro´ M, Ferrer A, et al. Multilevel Control of Arabidopsis 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase by Protein Phosphatase 2A. The Plant Cell. 2011; 23: 1494–1511.
- Enjuto M, Balcells L, Campos N, Caelles C, Arro´ M, Boronat A. Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc Natl Acad Sci USA. 1994; 91: 927–931.
- Enjuto M, Lumbreras V, Mari´n C, Boronat A. Expression of the Arabidopsis HMG2 gene, encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase, is restricted to meristematic and floral tissues. Plant Cell. 1995; 7: 517–527.
- Lumbreras V, Campos N, Boronat A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J. 1995; 8: 541–549.
- Pfaffl MW, Hageleit M. Validities of mRNA quantification using recombinant RNA and recombinant DNA external calibration curves in real-time RT-PCR. Biotechn Lett. 2001; 23: 275-282.
- Brito FF, Gimeno PM, Carnes J, Martin R, Fernandez-Caldas E, Lara P. et al. Olea europaea pollen counts and aeroallergen levels predict clinical symptoms in patients allergic to olive pollen. Ann. Allergy Asthma Immunol. 2011; 106: 146-152.
- Davies JM, Li H, Green M, Towers M, Upham JW. Subtropical grass pollen allergens are important for allergic respiratory diseases in subtropical regions. Clin Transl Allergy. 2012; 2: 1-10.
- Žiarovská J, Labajová M, Ražná K, Bežo M, štefúnová V, Shevtsova T. et al. Changes in expression of BetV1 allergen of silver birch pollen in urbanized area of Ukraine. J Env Sci Health. 2013; 48: 1479-1484.
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25: 3389–3402.
- Breiteneder H, Ferreira F, Hoffmann-Sommergruber K, Ebner C, Breitenbach M, Rumpold H. et al. Four recombinant isoforms of Cor a I, the major allergen of hazel pollen, show different IgE-binding properties. Eur J Biochem. 1993; 212: 355–362.
- Ebner C, Ferreira F, Hoffmann K, Hirschwehr R, Schenk S, Szépfalusi Z. et al. T cell clones specific for Bet v I the major birch pollen allergen cross-react with the major allergens of hazel Cor a I and alder Aln g I. Mol Immunol. 1993; 30: 1323–1329.
- Bryce M, Drews O, Schenk MF, Menzel A, Estrella N, Weichenmeier I. et al. Impact of urbanization on the proteome of birch pollen and its chemotactic activity on human granulocytes. Int Arch Allergy Immunol. 2010; 151: 46–55.
- Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 2008; 8: 131.
- Boava LP, Laia ML, Jacob TR, Dabbas KM, Goncalves JF, Ferro JA. Et al. Selection of endogenous genes for gene expression studies in Eucalyptus under biotic (Puccinia psidii) and abiotic (acibenzolar-S-methyl) stresses using RT-qPCR. BMC Res Notes. 2010; 3: 43.
- Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol. 2003; 52: 403–408.
- Cortleven A, Remans T, Brenner WG, Valcke R. Selection of plastid- and nuclear-encoded reference genes to study the effect of altered endogenous cytokinin content on photosynthesis genes in Nicotiana tabacum. Photosynth Res. 2009; 102: 21-29.
- Hu R, C Fan, H Li, Q Zhang, YF Fu. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RTPCR. BMC. Mol. Biol. 2009; 10: 93.
- DeSantis C, Smith-Keune C, Jerry DR. Normalizing RT-qPCR data: Are we getting the right answers? An appraisal of normalization approaches and internal reference genes from a case study in the finfish lates calcarifel. Mar Biotechnol. 2011; 13: 170-80.
- Jarošová J, Kundu JK. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010; 10: 146.