
Perspective
Austin J Genet Genomic Res. 2015; 2(2): 1013.
Gene Duplications and Fates of Genes after Duplication Events
Abhishek Kumar
¹Division of Molecular Genetic Epidemiology, German Cancer Research Center, Germany
²Department of Genetics & Molecular Biology in Botany, Christian-Albrechts-University at Kiel, Germany
*Corresponding author: Abhishek Kumar, Molecular Genetic Epidemiology, German Cancer Research Center (DKFZ), ImNeuenheimer Feld 580, 69120 Heidelberg, Germany
Received: September 03, 2015; Accepted: September 14, 2015; Published: September 15, 2015
Perspective
Gene duplications are considered to be major genetic basis for producing novel genetic variations. There are three types of gene duplications: Whole Genome (WGD), segmental and small scale duplications by tandem duplications [1]. Example of gene duplications of gene duplications events are frequently found in vertebrate serpins, which are classified into six groups V1-V6 [2]. Several tandem duplications on the same locus lead into several paralogs for groups V1 on the human chromosomes 6 and 18 [3] and V2 on the human chromosome 14 [4]. In contrast, there are several serpins which are localized on a single gene in the chromosomal fragments like angiotensinogen [chromosomes 1[5]], heparin cofactor II [chromosomes 22 [6]], C1 inhibitor [chromosomes 22 [7]] and antithrombin III [chromosomes 1 [8]], respectively. Similar trends are also followed by serpins from invertebrates like urochordates [9].
About five decades ago, Susumu Ohno proposed his famous hypothesis that early vertebrates have undergone two rounds of WGD events and which is known as Ohno’s hypothesis or 2R-hypothesis [10,11]. These events cause massive gene duplications that are the hallmarks of gene and functional innovation. There were not one who believed on this hypothesis in the beginning years; later same theory become the cornerstone of gene duplications and fates [10,11], specially in the post-genomic era [12]. It is now clear that second WGD has also occurred in fishes and this often called as Fish- Specific Genome Duplication (FSGD) or 3R-hypothesis and it is best exemplified in the Hox clusters duplications [13].
On the evolutionary scale, gene duplication events lead into several schemes (Figure 1).
This includes following schemes:
a) Nonfunctionalization is random loss of function in one of the two gene copies by pseudogenization (Figure 1A). Large fraction of duplicated genes are pseudogenized over 100 MY in the rainbow trout [14].
b) Neofunctionalization is when one gene copy may retain the original function while the other acquires a novel, evolutionarily advantageous/adaptive function [15].
c) Subfunctionalization is after duplication, mutations may occur in both genes that specialize to perform complementary functions [16,17].
There are several puzzles about how duplicate genes are retained during evolution. Several models have been proposed over in last four decades. Classical duplication-degeneration-complementation/ subfunctionalization models do not invoke positive selection; however, this can impose higher rates for retaining duplicate genes in small populations only. Rodents have higher retentions rates duplicate genes and only few losses in comparison to humans which corroborates that positive selections are more instrumental players than previously assumed [18].
Suppose a condition where two redundant gene copies were retained in the genome without significant functional divergence, this can covenant increase genetic robustness against harmful mutations in the concerned species (Figure 1C). Within multigene families descended from a common ancestor, these genes possess similarities at the DNA level, which implies for similar functions [19,20]. The tandemly duplicated genes which generate several paralogous on the same chromosomal fragments (like serpin paralogous in the human chromosomes 6-18 [3] and 14 [4]), exhibit the case of concerted evolution. Within this concept, all genes in a given group evolve coordinately by homologous recombination, which vanguard into gene conversion (Figure 1D). These paralogs share higher sequence identities like anti-trypsin-like gene cluster in the human chromosome 14 [4]. For large fraction of multigene families, the evolutionary model of birth-and-death (aka gain-and-loss) is largely supporting model, which propound that protein sequence similarities within family members is pronounced by strong purifying selection and evolvements of individual genes are primarily occur only by synonymous substitutions (Figure 1E) [19,20].
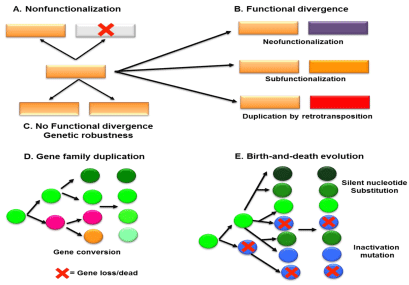
Figure 1: Fate of duplicated single genes (A-C) and duplicated gene families
(D-E).
Modified from Conrad and Antonarakis [1].
Vertebrate OVO-like transcription factors family depicts a good example of birth-and-death model of gene evolution (Figure 2). This family has lineage specific birth and dead of genes as reported earlier [21]. Birds and lizards have loss of OVOL1 and its entire locus, while ray-finned fishes have loss of entire OVOL2 locus. Fishes have duplication of OVOL3, generating original OVOL3a and duplicated OVOL3b. Subsequently fishes lost OVOL3a and OVOL3b has compensated for this loss.
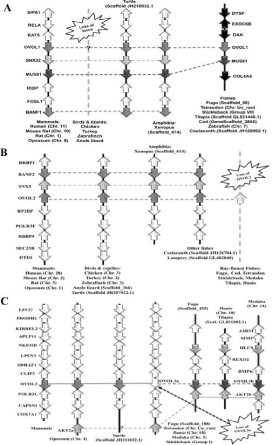
Figure 2: Birth-and-death model of gene evolution by example of vertebrate
OVO-like transcription factors family.
A) Loss of OVOL1 and its locus in birds and lizards.
B) Loss of OVOL2 in ray-finned fishes
C) Loss of original OVOL3a gene, which is compensated by duplicated
OVOL3b gene in ray-finned fishes.
Modified from Kumar et al. [21].
For further details of models of gene duplications and their fates, it is recommended to see a good review by Innan and Kondrashov [22] on this topic.
References
- Conrad B, Antonarakis SE. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet. 2007; 8: 17-35.
- Kumar A. Bayesian phylogeny analysis of vertebrate serpins illustrates evolutionary conservation of the intron and indels based six groups classification system from lampreys for ~500 MY. PEERJ. 2015:1-2.
- Benarafa C, Remold-O'Donnell E. The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proc Natl Acad Sci U S A. 2005; 102: 11367-11372.
- Forsyth S, Horvath A, Coughlin P. A review and comparison of the murine alpha1-antitrypsin and alpha1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003; 81: 336-345.
- Kumar A, Sarde SJ, Bhandari A. Revising angiotensinogen from phylogenetic and genetic variants perspectives. Biochem Biophys Res Commun. 2014; 446: 504-518.
- Kumar A, Bhandari A, Sarde SJ, Goswami C. Genetic variants and evolutionary analyses of heparin cofactor II. Immunobiology. 2014; 219: 713-728.
- Kumar A, Bhandari A, Sarde SJ, Goswami C. Molecular phylogeny of C1 inhibitor depicts two immunoglobulin-like domains fusion in fishes and ray-finned fishes specific intron insertion after separation from zebrafish. Biochemical and Biophysical Research Communications. 2014; 450: 219-226.
- Kumar A, Bhandari A, Sarde SJ, Goswami C. Sequence, phylogenetic and variant analyses of antithrombin III. Biochem Biophys Res Commun. 2013; 440: 714-724.
- Kumar A, Bhandari A. Urochordate serpins are Classified into Six Groups Encoded by Exon-Intron Structures, Microsynteny and Bayesian Phylogenetic Analyses. J Genomics. 2014; 2: 131-140.
- Ohno S. Evolution by Gene Duplication. New York: NY: Springer Verlag. 1970.
- Ohno S. Gene duplication and the uniqueness of vertebrate genomes circa 1970-1999. Semin Cell Dev Biol. 1999; 10: 517-522.
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999; 151: 1531-1545.
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000; 290: 1151-1155.
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000; 154: 459-473.
- Shiu SH, Byrnes JK, Pan R, Zhang P, Li WH. Role of positive selection in the retention of duplicate genes in mammalian genomes. Proc Natl Acad Sci USA. 2006; 103: 2232-2236.
- Nei M, Rogozin IB, Piontkivska H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc Natl Acad Sci USA. 2000; 97: 10866-10871.
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005; 39: 121-152.
- Kumar A, Bhandari A, Sinha R, Sardar P, Sushma M, Goyal P, et al. Molecular phylogeny of OVOL genes illustrates a conserved C2H2 zinc finger domain coupled by hypervariable unstructured regions. PLoS One. 2012; 7: e39399.