Abstract
Non-Alcoholic Fatty Liver Disease (NAFLD) is among the commonest causes of liver disease in the United States. Its progression to Non-Alcohol Steatohepatitis (NASH) increases risk for developing cirrhosis and liver cancer. Hepatocellular accumulation of triglycerides and cholesterol, the main lipids associated with NAFLD, is considered benign. In contrast, aberrant expression of sphingolipids and phospholipids that have structural and functional roles in cell membrane integrity and intra-cellular signaling, may mediate progression of NAFLD to NASH. This study utilized an established experimental model of NAFLD gen-erated after 16 weeks of High Fat Diet (HFD) feeding of adult (8 weeks old) male C57BL/6 mice. Fresh frozen liver tissue samples were used for lipidomics analysis by matrix-assisted laser desorption ioniza-tion-imaging mass spectrometry in the negative and positive ion modes. In HFD fed mice, histopatholog-ical changes of NAFLD were associated with pronounced alterations in hepatic lipid profiles marked by increased expression of phosphatidylcholines (54%), phosphatidylinositols (50%), phosphatidylglycerols (50%), and phosphatidylinositol monomannosides (100%); sphingolipids including ceramides (63%), sphingomyelins (54%), sulfatides (57%), mannose inositol phosphoceramides (100%), and glycosphin-golipids (50%); and glycerolipids including triacylglycerols (56%). In addition, NAFLD was associated with increased levels of hepatic arachidonic acid containing phosphatidylserine, phosphatidylinositol, and phosphatidylethanolamine species and depletion of docosahexaenoic acid containing phosphatidylserine. Correspondingly, PCA plots sharply distinguished between the HFD and low-fat diet control groups. Experimental NAFLD is associated with a broad array of increased hepatic lipids expression. These results establish a platform for evaluating mechanisms and consequences of hepatic lipidomic abnormalities that occur with progression from of NAFLD to NASH.
Keywords: Non-alcoholic Fatty Liver Disease; Steatohepatitis; Lipidomics; Mass Spectrometry; High Fat Diet; Mouse Model
Abbreviations
CER: Ceramide; DHB: Diaminobenzidine; ER: Endoplasmic Reticulum; GSL: Glycosphingolipids; HBSS: Hanks Balanced Salt Solution; HCCA: a-Cyano-4-hydroxycinnamic Acid; HFD: High Fat Diet; IMS: Imaging Mass Spectrometry; ITO: Indium Tin Oxide; LFD: Low Fat Diet; m/z: Mass-to-Charge ratio; MALDI: Matrix- Assisted Laser Desorption/ionization; MS: Mass Spectrometry; NAFLD: Non-Alcoholic Fatty Liver Disease; NASH: Non Alcoholic Steatohepatitis; OCT: Optimal Cutting Temperature Compound; PA: Phosphatidic Acids; PC: Phosphatidylcholines; PCA: Principal Component Analysis; PE: Phosphatidylethanolamines; PG: Phosphatidylglycerols; PI: Phosphatidylinositols; PIM: Phosphatidylinositol Monomannosides; PS: Phosphatidylserines; SM: Sphingomyelin; TG: Triglycerides; TMA: Tissue Microarray; TNF: Tumor Necrosis Factor; TOF: Time of Flight
Introduction
Nonalcoholic Fatty Liver Disease (NAFLD) encompasses a broad spectrum of pathological states that begin with benign or simple steatosis, but in a subset of individuals, leads to Non-Alcoholic Steatohep-Atitis (NASH) with eventual progressive development of fibrosis, cirrhosis, and finally end-stage liver disease [1,2]. NAFLD is linked to metabolic risk factors such as obesity, type 2 diabetes mellitus, insulin resistance, and cardiovascular disease [1]. Current estimates are that in the United States, approximately 64 million people have NAFLD, many of whom have not yet been diagnosed [3]. The burdens posed on quality of life and personal as well as healthcare economics continue to grow, in part due to presently limited effective therapeutic options [4].
The pathogenic mechanisms of NAFLD development and progression of are linked to a wide range of cellular and molecular pathologies including insulin resistance, metabolic derangements altering lipid metabolism, inflammation, oxidative stress, DNA damage, and mitochondrial dysfunction [5-7]. However, the main drivers of this cascade are insulin resistance through metabolic pathways and dysregu-lated lipid metabolism. Dietary fat, sugars, adipose tissue lipolysis, and de novo lipogenesis increase he-patic lipid content [8,9]. Insulin resistance is permissive to lipolysis and negatively affects the ability of the adipose tissue to store fat resulting in increased free fatty acids in the blood [10,11]. Hepatic de novo lipogenesis is also augmented with metabolic syndrome due to insulin resistance and ER stress [12,13]. Lipid accumulation in the liver primarily consists of triglycerides, which may not be hepatotoxic and serve as a protective mechanism to prevent fatty acid mediated liver injury [14,15]. On the other hand, long chain saturated fatty acids have been shown to be elevated in NASH patients and cause injury in liv-er cells by triggering formation of reactive oxygen species and lipid peroxidation that contribute to hepat-ic lipotoxicity [11,16].
There is a growing recognition that multiple lipid classes are involved in the pathophysiology of NAFLD. Hence, the role of specific lipid classes, rather than total hepatic fat or triglyceride content, in the development and progression of NAFLD is emerging [17,18]. Previous studies conducted on experi-mental models of diet induced obesity with NAFLD showed increased hepatic ceramide levels through activation of de novo biosynthesis and sphingomyelin degradation pathways resulting in insulin re-sistance, lipotoxicity, ER/mitochondrial stress, and inflammation [7,19,20]. Along with fatty acids and ceramides, diacylglycerols play key roles in mediating inflammatory pathways leading to lipotoxicity and oxidative stress, thus contributing to NAFLD progression [21,22]. In addition, metabolic studies provid-ed insights into NAFLD associated alterations in phospholipid profiles. Phosphatidylcholine (PC) and Phosphatidylethanolamine (PE) are the two most abundant phospholipids in plasma membranes of all mammalian cells and a change in their absolute concentrations is a key determinant of liver health and disease. The circulating PC levels significantly increased [23], while hepatic PC content decreased [18] in NAFLD and NASH patients relative to healthy control subjects implicating an important pathophysiolog-ical role for this lipid class. Furthermore, relatively small alterations in hepatic PC/PE molar ratio can im-pair membrane integrity and contribute to the development of NAFLD [24,25].
Recent advancements in lipidomics analysis have enabled characterization of membrane phos-pholipids, sphingolipids, and glycerolipids and the study of altered membrane lipid profiles in relation to pathophysiological conditions [26,27]. Since NAFLD is defined by imbalances in lipid homeostasis, lip-idomics approaches are applicable to investigations of how lipid metabolism is altered with disease. Pre-vious studies mainly focused on ceramides yet the contributions of other sphingolipids such as sphingo-myelins and sulfatides, and the various subtypes of phospholipids are less wellknown . The goal of this study is to use an experimental mouse model of high fat diet induced NAFLD to characterize alterations in hepatic lipid profiles using MALDI imaging mass spectrometry.
Materials and Methods
Materials
HPLC grade solvents, 2,5-Dihydroxybenzoic Acid (DHB), a-Cyano-4-Hydroxycinnamic Acid (HCCA), polyvinyl alcohol 6-98, Polypropylene Glycol (average MW 2,000 g/mol (PPG 2000)), and sodium azide were purchased from Sigma Aldrich (St. Louis, MO). Hanks Balanced Salt Solution (HBSS) was purchased from Lonza (Allendale, NJ). Tissue Microarray (TMA) mold and coring tools were purchased from Arraymold Kit (Salt Lake City, UT). Peptide calibration standards were purchased from Bruker Daltonics (Billerica, MA). Male C57BL/6 mice were purchased from Jack-son Laboratories. High fat diet (F3282) was purchased from BioServ (Marlborough, MA).
Experimental Model: Eight-week-old C57BL/6 male mice (n=6 per group) were pair-fed with high fat or low fat (chow) diets for 16 weeks. The HFD consisted 60% kcal fat from lard, whereas the normal chow diet contained 18% kcal fat (Supplementary Table 4). Mice were housed under humane con-ditions with free access to food. Food intake was monitored daily and body weight was measured weekly. Mice were sacrificed by isoflurane inhalation and cervical dislocation and their livers were harvested im-mediately. Liver tissue samples were frozen on dry ice and stored at -80°C for later MALDIIMS analy-sis. This study was approved by the Institutional Animal Care and Use Committee at Lifespan-Rhode Is-land Hospital, and the experimental protocol followed the guidelines established by the National Institutes of Health.
MALDI-IMS
Frozen liver tissues were used to generate a TMA to enable simultaneous analysis of all samples under identical conditions. Frozen livers (n=6 per group) were cored using a 1.5-mm diam-eter Arraymold coring tool and transferred into a TMA mold made with modified OCT. TMA enabled simultaneous acquisition and analysis of all samples in a single imaging data set. Modified OCT was used as the embedding compound because it does not interfere with mass spectrometry signals [28]. Two con-secutive cryosections (8 μm thick) of the TMA block were mounted onto an Indium Tin Oxide (ITO)-coated slide side by side. 200 ± 13 mg/cm2 of DHB was applied onto the slide by sublimation as de-scribed previously [29]. One TMA section was imaged in the negative ion mode and the other one was imaged in the positive ion mode with an Ultraflextreme MALDItime- of-flight (TOF/TOF) mass spec-trometer (Bruker Daltonics, Billerica, MA). A Smartbeam II Nd:YAG laser, providing a laser focus down to 25 μm in diameter, was selected for the acquisition of imaging data, with a laser raster step size of 75 μm and 500 laser shots summed per array position (i.e., per pixel). External mass calibration was carried out in a cubic enhanced mode using matrix (HCCA) and peptide mixture to obtain at least five calibration points over the mass range between 377 and 2463 Da. Consecutive negative and positive ion mode IMS measurements were acquired from 600-1200 Da mass range in a reflectron mode. Ions were accelerated at 25 and 20 kV with 90 and 140 ns of pulsed ion extraction delay with the extraction voltage at 22 and 17 kV in the positive and negative ion modes, respectively.
Data Analysis
The pre-processing of MALDI imaging data was performed by normalization of all mass spectra to Total Ion Count (TIC) with FlexImaging software version 4.0 (Bruker Daltonics). TIC is a standard normalization method where all mass spectra are divided by their TIC (the sum of all intensi-ties) to enable all spectra in a dataset to have the same integrated area under the spectrum [30]. The com-plete MALDI-TOF MS spectra obtained from each sample within the TMA (90 spectra per sample) was imported into ClinProTools software for post-processing including the generation of lipidomic profiles. Normalizing, baseline subtracting, peak defining, recalibrating, and comparison of multiple spectra were performed automatically by the Clin ProTools software. Tentative lipid assignment was made by compar-ing mass-to-charge (m/z) values of precursor ions with previously identified lipids in our laboratory or other published reports. The average intensity of lipid ions per group was used to compare HFD mediated alterations relative to LFD samples. Data bar plots were used to visualize the mean percent changes in lipid ion expression. Inter-group comparisons were made by T-tests with a 5% false discovery (GraphPad Prism 8, San Diego, CA, USA). Principle Component Analysis (PCA) generated in ClinProTools was used to compare lipid ion expression patterns between HFD and LFD. Chi-square analysis with Yates’ correc-tion (GraphPad Prism 8, San Diego, CA, USA) was used to determine whether HFD differentially altered expression of major lipid classes (phospholipids, sphingolipids, and glycerolipids) and their subclasses. P-values less than 0.05 were considered as statistically significant.
Results
Hepatic Lipid Profiles
The peak statistics reports obtained from Clin ProTools identified 241 ions in the negative ion mode and 151 ions in the positive ion mode between the mass-to-charge ratio of 600 and 1200 Da. Putative lipid annotations made by previous identifications performed in our laborato-ry, published literature [31-52], or Lipid Maps database search (http://www.lipidmaps.org/tools/ms/) and are listed in Supplementary Table 3. The lipids detected in the positive ionization mode formed in proton ([M+H]+), sodium ([M+Na]+), potassium ([M+K]+), and ammonium ([M+NH4]+) adducts, while the nega-tive ionization mode formed only deprotonated adducts ([M-H]-). Lipid classes include 1) phospholipids (n=170; 43.4%), including 54 (13.8%) Phosphatidylcholines (PCs), 19 (4.8%) Phosphatidylethanolamines (PEs), 21 (5.4%) Phosphatidylserines (PSs), 43 (11%) Phosphatidylinositols (PIs), 4 (1%) Phosphatidyl-Glycerols (PGs), 6 (1.5%) Phosphatidic Acids (PAs), 2 (0.5%) Phosphatidylinositol Monomannosides (PIMs), and 21 (5.4%) Phospholipids (head group unidentified); 2) sphingolipids (n=81; 20.7%), includ-ing 50 (12.8%) Sphingomyelins (SMs), 15 (3.8%) Sulfatides (STs), 4 (1%) Ceramides (CERs), 3 (0.8%) Hexosylceramides (HexCers), and 4 (1%) Lactosylceramides (LacCers), 2 (0.5%) Mannose Inositol Phos- Phoceramides (MIPCs), and 3 (0.8%) Glycosphingolipids (GSLs); 3) Glycerolipids (n=56; 14.3%), includ-ing 43 (11%) Triacylglycerols (TGs) and 13 (3.3%) Diacylglycerols (DGs); and 4) miscellaneous ions (head group unidentified) (n=24; 6.1%) or unidentified (n=61; 15.6%) (Supplementary Table 1).
TABLECREATED
Table 1: Effects of HFD on lipid classes expressed in mouse livers.
Differential Expression of Lipid Ions in HFD- and LFDExposed Mouse Livers
To optimize the comparison of lipid ion expression in HFD and control samples, 6 standardized paired samples from each group were put into a TMA block and sectioned on an ITO coated slide for simultaneous imaging under identical conditions for data acquisition. The hepatic lipid composition of LFD and HFD fed mice were similar with 290 ions expressed in both groups. However, livers of HFD fed mice expressed 24 li-pids that were not detected in the livers of LFD fed mice, and LFD fed mouse livers expressed 39 lipids that were not detected in HFD fed mouse livers (Supplementary Table 2). HFD mouse livers differentially expressed 13 phospholipids, 4 sphingolipids, 10 glycerolipids, and 21 unidentified lipids that were not observed in LFD fed controls. LFD livers expressed 12 phospholipids, 8 sphingolipids, 15 glycerolipids, and 17 unidentified lipids that were not observed in HFD mouse livers. Chi-square analysis with Yates’ correction determined that the differential expression of phospholipids (X2=21.15, 1df; P<0.0001), sphingolipids (X2=7.92, 1df; P=0.005), glycerolipids (X2=21.01, 1df; P< 0.0001), and unidentified lipids (X2 =34.06, 1df; P< 0.0001) were statistically significant. These differentially expressed lipids demonstrate that the lipid compositions of LFD and HFD mouse livers were not identical.
HFD Effects on hepatic lipid profiles determined by Principle Component Analysis (PCA)
PCA was used to reduce the dimensionality of the MALDIIMS data set, while retaining the in-formation present within the mass range of 600 and 1200 Da. PCA plots from positive ion mode (Figures 1A-D) and negative ion mode (Figures 1E-H) data sets including the full spectra acquired between 600 and 1200 Da show the distinction between HFD and LFD fed mouse liver lipid profiles. The 3-dimensional (Figures 1A and 1E) and 2-dimensional (Figures 1B and 1F) PCA plots demonstrated clear separation of the groups on the PC1×PC2 plane. Although some data points from HFD and LFD do not show much varia-tion on PC1 x PC3 (Figures 1C and 1G) and PC2×PC3 (Figures 1D and 1H) planes, their separation was confirmed in the 3-D plots. The separated clusters of two experimental groups represent differential effects of HFD on hepatic lipid ion profiles.
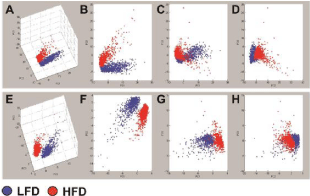
Figure 1: Principal Component Analysis (PCA) of hepatic lipids detected
by MALDI-IMS in the (A-D) positive and (E-H) negative ionization modes.
MALDI-IMS spectra acquired between 600 and 1200 Da mass range were
compared between HFD fed (red) mouse livers and LFD controls (blue).
PCA plots were generated in ClinProTools. Panels A and E represent the
3-dimensional PCA plots. B–D and F–H correspond to 2-dimensional
plots of A and E, respectively. Note the clear separation of HFD and LFD
clusters in 3 dimensional plots. The contributions of PC1, PC2, and PC3 were
approximately 50%, 12%, and 10%, respectively.
HFD Effects on hepatic lipid expression demonstrated by data bar plots
Comparative lipid analysis of the livers of HFD and LFD (n=6 per group) fed mice revealed relative effects of HFD on phospholipids, sphingolipids, and glycerolipids detected between a 600-1200 Da mass range in positive and negative ion modes. Data presentation was simplified by including only the lipids detected in both control and HFD samples. To evaluate the effects of HFD on lipid expression, the mean peak intensities (reflecting lipid abundance) were compared by t-test analysis with a 5% false discovery rate correction. The percent change differences in mean lipid ion abundance were graphed in data bar plots to visualize relative effects of HFD on hepatic lipid ion expression. Data bar plots were separated by lipid subclass and lipids were detailed in the ascending order based on the total number of carbon atoms and double bonds in the structure. HFD-associated reductions in lipid ion expression were represented by the blue bars to the left, whereas HFD-associated increases in lipid expression were indicated by red bars to the right (Figures 2-4). The differences lower than 5% were considered as unchanged. Significant (p< 0.05) or trend (0.05< p< 0.1) effects obtained by t-test analysis were shown next to the corresponding data bars. The overall effects of HFD on the expression levels of different lipid classes are summarized in Table 1.
Phospholipids: The phospholipid bar plots included 50 Phosphatidylcholines (PCs), 14 Phospha-Tidylethanolamines (PEs), 17 Phosphatidylserines (PSs), 40 Phosphatidylinositols (PIs), 4 Phosphatidyl-Glycerols (PGs), 3 Phosphatidic Acids (PAs), 2 Phosphatidylinositol Monomannosides (PIMs), and 11 Phospholipids (PLs) (Figure. 2). HFD increased expression of 72 (51.1%) phospholipid ions, reduced expres-sion of 55 (39%) ions, and had no effect on 14 (9.9%) ions, relative to LFD fed controls (Table 1).

Figure 2A-2C: Data bar plots demonstrating effects of HFD on hepatic
phospholipid ion expression as detected by MALDI-IMS in the positive and
negative ion modes. Plots represent percent change differ-ences in mean
levels of A) phosphatidylcholines (PCs), B) phosphatidylethanolamines
(PEs), C) phospha-tidylserines (PSs).
Phosphatidylcholines were the most abundant phospholipids detected as proton, sodium, or potas-sium adducts in positive ion mode in the mouse livers (Figure 2A). HFD had dramatic effects on PC expres-sion that ranged from a 58% decrease to a 167% increase. Among 50 PC species expressed both in LFD and HFD mouse livers, 27 PCs (54%) were expressed at higher levels and 18 (36%) were expressed at lower levels in HFD livers relative to control, while 5 PCs (10%) had no change with HFD feeding (Table 1). HFD mediated changes were statistically significant for 23 PCs and trend effects were observed for 13 PCs, whereas the remaining 14 PCs failed to reach significance due to low levels of percent difference between HFD and LFD livers (Figure 2A).
Phosphatidylethanolamines were detected in both positive and negative ion modes with half of them ionized as deprotonated adducts, 6 PEs as protonated adducts, and 1 PE as a sodium adduct (Figure 2B). Within 14 PE ions, 12 of them were putatively annotated as parent ions and 2 were C13 isotopes. HFD increased hepatic expression of 6 PEs (42.9%), reduced 6 PEs (42.9%), and had no effect on 2 PEs (14.3%) (Table 1). These alterations were statistically significant except for 4 PE species that showed less than 10% change with HFD feeding.
Phosphatidylserine species were detected exclusively in the negative ion mode as deprotonated adducts. HFD increased hepatic expression of 7 PSs (41.2%), reduced 8 PSs (47.1%), and had no effect on 2 PSs (11.8%) (Table 1). HFD-induced increases or reductions were statistically significant for 11 PS ions and trend effects were observed for 3 PS ions (Figure 2C). It is noteworthy that hepatic expression of docosahexaenoic acid containing PS, PS (40:6), was significantly decreased (30%, P=0.01) in HFD-exposed livers relative to controls. In contrast, arachidonic acid containing PS, PS(38:4), and its C13 isotope were increased (70% and 52%, respectively, P<0.0001) by HFD feeding.
Phosphatidylinositols were also detected in the negative ion mode as deprotonated adducts and putatively annotated as oxidized (OS-PI), Phosphorylated (PIP), or unconjugated forms (PI). Among 40 ions, HFD increased 20 PIs (50%), decreased 17 PIs (42.5%), and had no change on 3 PIs (7.5%) (Table 1). T-test analysis revealed that the alterations in 16 of 20 PIs expressed at higher levels and all 17 PIs expressed at lower levels in HFD livers were statistically significant or had trend effects (Figure 2D). In ad-dition, hepatic expression of arachidonic acid containing PI, PI(38:4), increased (78%, P<0.0001) in HFD fed mouse livers relative to controls. In contrast, HFD had a trend reduction effect on oxidized PI expres-sion relative to the control (-47%, P=0.07).
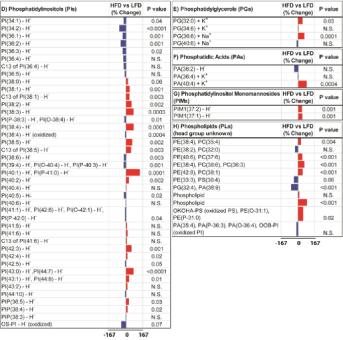
Figure 2D-2H: D) phosphatidylinositols (PIs), E) phosphatidylglycerols
(PGs), F) phosphatidic acids (PAs), G) phosphatidylinositol monomannosides
(PIMs), and H) phospholipids (PLs) with head groups unidentified. The scale
bars depict HFD mediated responses ranged between -167% and 167%
relative to controls. P values obtained by T-test analysis of comparing the
mean levels of each phospholipid ion in LFD and HFD groups are shown
to the right of each data bar plot. Results are organized with respect to
increasing total number of carbon atoms and double bonds of phospholipids.
HFD mediated reductions in phospholipid expression are represented by
the blue bars to the left of the vertical axis, and increases by the red bars
to the right correspond to mean percentage increases in phospholipid ion
expression.
Phosphatidylglycerols and phosphatidic acids expressed fewer species than other phospholipids, only 4 PG and 3 PA species were detected in mouse livers. HFD caused an increase in hepatic expression of PG(32:0) (44%, P=0.03), PG(36:6) (61%, P=0.0001), and PA(40:4) (85%, P=0.0004), while the other 2 PGs and 2 PAs did not change significantly (Figures 2E,2F).
Two Phosphatidylinositol Monomannosides, PIM1(37:1) and PIM1(37:2), were detected in the negative ion mode. HFD increased hepatic expression of both PIMs by 37% and 39% relative to control samples (P=0.001) (Figure 2G).
We detected 11 phospholipids with head groups that could not be further identified. HFD in-creased 7 PLs (63.6%) and decreased 4 PLs (36.4%) (Table 1). T-test analyses showed significant differ-ences for 6 PLs expressed at higher levels and for only 1 PL expressed at a lower level in HFD-exposed livers relative to controls (Figure 2H). Another cluster of lipids containing 19 ions were assigned as phos-pholipids or sphingolipids. Similar to phospholipids, HFD increased 13 lipids and decreased only 1 lipid significantly. In addition, 30 ions remained unidentified after Lipid Maps and literature search (data not shown). HFD had similar effects on these unassigned peaks, significantly increased expression of 10 ions whereas it decreased only 1 ion in mouse liver samples.
Sphingolipids: The sphingolipid bar plots included 43 Sphingomyelins (SMs), 14 Sulfatides (STs), 10 Ceramides (CERs), and 2 Glycosphingolipids (GSLs) (Figure 3). HFD increased expression of 39 (56.5%) sphingolipid ions, reduced expression of 22 (31.9%) ions, and had no effect on 8 (11.6%) ions, relative to LFD fed controls (Table 1).
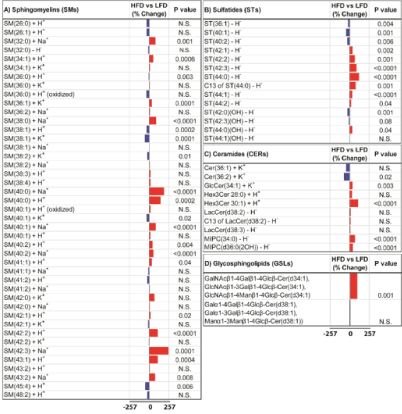
Figure 3: Data bar plots demonstrating effects of HFD on hepatic sphingolipid
ion expression as detected by MALDI-IMS in the positive and negative ion
modes. Plots represent percent change differ-ences in mean levels of A)
sphingomyelins (SMs), B) sulfatides (STs), C) ceramides (CERs), and D)
gly-cosphingolipids (GSLs). The scale bars depict HFD mediated responses
ranged between -257% and 257% relative to controls. P values obtained
by T-test analysis of comparing the mean levels of each sphin-golipid ion
in LFD and HFD groups are shown to the right of each data bar plot. Results
are organized with respect to increasing total number of carbon atoms and
double bonds of sphingolipids. HFD mediat-ed reductions in sphingolipid
expression are represented by the blue bars to the left of the vertical axis,
and increases by the red bars to the right correspond to mean percentage
increases in sphingolipid ion ex-pression.
Sphingomyelins (SMs) were mainly detected in the positive ion mode as sodium, potassium, or proton adducts except for SM(32:0) which ionized in deprotonated form. Sphingomyelin data bar plot revealed that HFD increased expression of 23 (53.5%) ions of the 43 ions detected, reduced expression of 14 (32.6%) ions, and had no effect on 6 (14%) ions (Table 1). HFD induced dramatic increases ranged between 50% to 257% and were observed for SM(34:1), SM(40:2), SM(42:0), SM(43:2), SM(32:0), SM (40:1), SM(38:0), SM(42:2), SM(43:1), SM(40:0), SM(40:0), and SM(42:3). In contrast, the degree of HFD-induced reductions in hepatic SM expression was lower ranging between -8 and -59%. T-test analy-sis revealed that increased hepatic expression of 15 SMs and reductions in 6 SM species were statistically significant (Figure 3A).
Sulfatides (STs) and Hydroxylated Sulfatides (ST-OHs) were detected as deprotonated adducts in the negative ion mode. HFD increased hepatic expression of 7 STs and 1 ST-OH, reduced 3 STs and 2 ST -OH, and had no effect on 1 ST-OH ion. T-test analysis showed that HFD-associated changes in hepatic ST levels were statistically significant (Figure 3B).
Among the 10 ceramides detected in positive and negative ion modes as protonated, potassiated, or deprotonated forms, HFD significantly increased expression of GlcCer(34:1), Hex3Cer(28:0), Hex3Cer (30:1), MIPC(34:0), and MIPC(d36:0(2OH)), while it decreased Cer(36:2) expression in the liver. Lacto-syl ceramide ions including LacCer(38:3), LacCer(38:2), and the C13 isotope of LacCer(38:2) were ex-pressed at higher levels in HFD fed livers relative to LFD controls, but t-test analysis failed to reach sta-tistical significance (Figure 3C).
Two Glycosphingolipids (GSLs) were detected at m/z 1063 and 1078 Da in the negative ion mode. Putative annotation by Lipid Maps searching resulted in several potential isobaric glycosphin-golipid species (Figure 3D and Supplementary Table 3). HFD significantly increased expression of m/z 1063 relative to control but had no significant effect on the expression of 1078 in liver.
Glycerolipids: As glycerolipids, we detected 27 Triacylglycerols (TGs) and 4 Diacylglycerols (DGs) expressed in both HFD and LFD fed mouse livers. Since the lipidomics analysis was focused be-tween 600-1200 Da mass range, we didn’t detect low mass Monoacylglycerols. TG species were detected as proton, potassium, sodium, or ammonium adduct forms in the positive ion mode. HFD increased hepat-ic expression of more than half of the TG species with 9 TGs increased significantly and 5 TGs with a statistical trend effect (Figure 4A). HFD reduced hepatic expression of 11 TGs, 5 of which were determined to be statistically significant. In contrast to TGs, HFD had mainly inhibitory effects on DGs (Figure 4B).
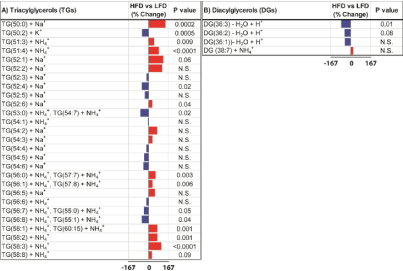
Figure 4: Data bar plots demonstrating effects of HFD on hepatic glycerolipid
ion expression as detected by MALDI-IMS in the positive ion mode. Plots
represent percent change differences in mean levels of A) triacylglycerols
(TGs) and B) diacylglycerols (DGs). The scale bars depict HFD mediated
responses ranged between -167% and 167% relative to controls. P values
obtained by T-test analysis of comparing the mean levels of each glycerolipid
ion in LFD and HFD groups are shown to the right of each data bar plot.
Results are organized with respect to increasing total number of carbon atoms
and dou-ble bonds of glycerolipids. HFD mediated reductions in glycerolipid
expression are represented by the blue bars to the left of the vertical axis,
and increases by the red bars to the right correspond to mean per-centage
increases in glycerolipid ion expression.
Out of 4 DG species, 3 of them had lower expression and 1 had higher expression in HFD mouse livers relative to controls. The reduction in DG(36:3) expression was statistically significant.
HFD Effects on Hepatic Lipid Expression Analyzed by Chi Square Analysis: The overall HFD mediated changes (increased, decreased, or unchanged) in hepatic lipid subclasses were determined by Chi-square analysis (Figure 5). HFD increased the majority of phospholipids (51.1%), sphingolipids (56.5%), and glycerophospholipids (51.6%) (Table 1). Chi-square analysis revealed that these changes on hepatic lipids were statistically significant (X2 = 14.29, 4 df; P = 0.006) (Figure 5A). Regarding phospholip-ids, HFD increased PCs (54%), PIs (50%), PGs (50%), PIMs (100%), and head group unidentified PLs (64%); reduced most of the PSs (47%); and had equal effects on PEs (43%) and PAs (33%) (Table 1). Chi square analysis demonstrated that HFD mediated changes in hepatic expression of total phospholipid sub-classes were statistically significant (X2=45.65, 10 df; P < 0.0001) (Figure 5B). HFD dramatically in-creased expression of all sphingolipid classes, including CERs (63%), SMs (54%), STs (57%), MIPCs (100%), and GSLs (50%). Chi square analysis revealed significant overall (including all subtypes) effects of HFD on hepatic sphingolipid expression (X2=30.80, 6 df; P< 0.0001). Regarding glycerolipids, HFD increased the majority of TGs (55.6%) and decreased DGs (55.6%). Chi square analysis of HFD effects demonstrated significant difference in the relative responses on the total pool of TGs and DGs (X2=19.87, 2 df; P < 0.0001).
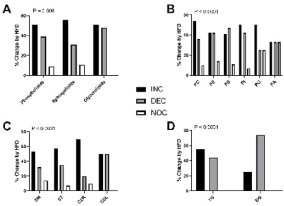
Figure 5: Combined effects of HFD-associated changes on hepatic lipid ion
expression. HFD’s overall effects on the hepatic expression of different lipid
subclasses relative to controls were evaluated statistically by Chi-square
analysis. The percentages of A) phospholipids, sphingolipids, and glycerolipids;
B) phosphatidylcholines (PCs), phosphatidylethanolamines (PEs),
phosphatidylserines (PSs), phosphatidylinositols (PIs), phosphatidylglycerols
(PGs), and phosphatidic acids (PAs); C) sphingomye-lins (SMs), sulfatides
(STs), ceramides (CERs), and glycosphingolipids (GSLs); and D)
triacylglycerols (TGs) and diacylglycerols (DGs) that were increased (INC),
decreased (DEC), or unchanged (NOC, <5% change) in HFD fed mouse
livers relative to control samples are shown on the graphs. P values obtained
from Chi-square tests are shown in the panels.
Discussion
This study utilized a chronic High Fat Diet (HFD) mouse model to mimic diet induced Non-Alcoholic Fatty Liver Disease (NAFLD), which is considered tobe a hepatic manifestation of metabolic syndrome. To investigate HFD mediated hepatic lipid abnormalities in NAFLD, we used C57BL6 because this strain is widely used in diabetes research and is highly susceptible to the development of insulin resistance and type II diabetes when placed on a HFD [53]. In our model, HFD feeding for 16 weeks result-ed in increased macro-vesicular and micro-vesicular steatosis with disorganization of hepatic chord archi-tecture as demonstrated by histopathological examination of Hematoxylin and Eosin stained livers (Supplementary Figure 1). Although previous studies linked NAFLD with accumulation of triglycerides as well as toxic fatty acids and ceramides, global changes in the hepatic lipidome including specific types and amounts of lipids have not been extensively investigated. The goal of this study was to perform a comprehensive lipidomics analysis to characterize HFD mediated alterations in hepatic lipid profiles in-cluding subclasses of phospholipids, sphingolipids, and glycerolipids and identify clustered responses to chronic HFD in an experimental model of NAFLD.
The composition of hepatic lipids detected by MALDI-IMS was similar to previous reports [18,47,54]. These include phospholipids consisting of Phosphatidylcholines (PCs), Phosphatidylethanola- Mines (PEs), Phosphatidylserines (PSs), Phosphatidylinositols (PIs), Phosphatidylglycerols (PGs), Phos-Phatidic Acids (PAs), and Phosphatidylinositol Monomannosides (PIMs), Sphingolipids Including Sphingo-Myelins (SMs), Sulfatides (STs), Ceramides (CERs), and Glycosphingolipids (GSLs), and Glycerolipids Comprised Of Triacylglycerols (TGs) and Diacylglycerols (DGs). In addition, fatty acids, cholesterol, and monoacylglycerol species that are abundantly expressed in liver were not detected in this study due to their relatively small mass (<600 Da).
Chronic HFD feeding caused distinct features in hepatic lipid composition and profiles as demon-strated by clearly separated clusters of HFD and control groups in PCA plots and lipids exclusively ex-pressed in HFD or control livers. The magnitude and direction of responses to HFD were revealed by data bar plots. The overall conclusion drawn from data bar plots was that HFD caused prominent increases in the hepatic expression of the majority of phospholipids, sphingolipids, and glycerolipids, while modest reductions occurred in fewer lipid sub-clusters. The dramatic increases or moderate declines in hepatic lipids could have reflected 1) alterations in membrane integrity and permeability mainly due to impaired phospholipid homeostasis, 2) increased insulin resistance and inflammation in response to accumulation of toxic sphingolipids, or 3) impaired cellular signaling functions that mediate proliferation, survival, and apoptosis.
Recent studies provided evidence for altered phospholipid metabolism in NAFLD, implicating an important pathophysiological role for this lipid class. PCs and PEs are the major structural components of the plasma membrane and are important sources of DGs and fatty acid derived secondary messengers [55]. The PC/ PE ratio is a critical modulator of membrane integrity and plays a key role in the progres-sion of NAFLD [24]. Several studies reported increased concentration of PCs and PEs in the systemic circulation of NAFL and NASH patients [23,56], whereas Puri et al. showed a significant decrease of PC and PE levels in the livers of NAFLD patients [18]. Our results from MALDI-IMS analysis of mouse livers demonstrated that hepatic expression of the majority of PCs (54%) was elevated by HFD feeding, while differing effects were observed for PE species (43% increase and 43% decrease). Inconsistent re-sults in the literature could be attributed to the sensitivity of the detection methods. Puri et al. used thin layer chromatography to separate phospholipids, whereas we utilized mass spectrometry analysis that pro-vides a better sensitivity and accuracy. HFD-associated reductions in hepatic PC and PE content could negatively impact hepatic membrane integrity and the permeability required for cell survival, growth, and proliferation. Conceivably, HFD-associated increases in PC and PE species may reflect compensatory responses linked to hepatocyte proliferation and regeneration.
With high throughput lipidomics analysis, there is a growing recognition that a multitude of phos-pholipids are potentially involved in the pathogenesis of chronic liver disease. In addition to PC and PE, we found HFD mediated alterations in PS and PI. These phospholipids are associated with inflammation and cellular apoptosis [57,58] and could therefore be related to the severity and progression of NAFLD. About half of the PS and PI species were significantly higher in livers of HFD fed mice, whereas the other half was lower relative to controls. Other studies reported increased levels of circulating PS and PI in NAFL and NASH patients [56,59]. PS has an important role in membrane stability and cellular apopto-sis. Typically, it is found in the inner cytosolic membrane and participates in intracellular signal transduc-tion [60]. However, PS is externalized to the cell surface in response to stimuli signaling for cell death. PI and its phosphorylated metabolites are second messengers involved in Mitogen Activated Protein Kinase (MAPK) and Protein Kinase B (PKB)/Akt signaling pathways [61]. In addition, PIs regulate vesicular traf-ficking, modulate lipid distribution and trafficking via lipid transfer proteins, and control membrane fluid-ity and permeability [61]. Conceivably, HFD-mediated alterations in hepatic PS and PI content could im-pair a number of physiological functions including PI3 kinase activation of Akt pathways, membrane sta-bilization, and apoptosis.
Analysis of the fatty acid composition of phospholipids revealed that HFD feeding significantly DECREASED Docosahexaenoic Acid (DHA) containing PS, PS(40:6). Based on their anti-inflammatory and anti-oxidant properties, growing evidence supports the therapeutic potential of omega-3 fatty acids, main-ly DHA, on metabolic diseases [62,63]. Mechanistic studies demonstrated that DHA containing PS suppressed hepatic SREBP-1 mediated lipogenesis and activated PPARa mediated fatty acid β-oxidation in the liver [64]. In addition, arachidonic acid containing PS, PI and PE were significantly increased by HFD. Arachidonic acid is liberated by phospholipase A2 for the production of eicosanoids that are in-volved in inflammatory processes [65]. These studies suggest that HFD mediated increases in arachidonic acid containing phospholipids and reductions in DHA containing PS may contribute to inflammation and oxidative stress in the liver.
Sphingolipids are bioactive lipids involved in regulating major biological functions including cell survival, proliferation, apoptosis, differentiation, migration, and immune responses [66,67]. The role of sphingolipids, especially ceramides, as mediators of insulin resistance and hepatotoxicity has been demonstrated in experimental models of chronic HFD feeding [7,72-75]. Our findings are in agreement with previous reports showing that HFD feeding resulted in increased levels of sphingolipids including ceramides (62.5%), sulfatides (57.1%), and sphingomyelins (53.5%). Increased ceramide species reflect the activation of the de novo ceramide synthesis pathway. Furthermore, degradation of sphingomyelins and sulfatides yields ceramides that promote insulin resistance, inflammation, formation of reactive oxy-gen species, and apoptosis [70,71]. In the present study, increased expression of ceramide may have re-sulted mainly from de novo synthesis since only 32.6% of SMs were decreased after chronic HFD expo-sure. The prominent upregulation of ceramides may have contributed to hepatic insulin resistance and injury as observed previously with chronic HFD feeding.
The HFD-induced upregulation of hepatic SMs is consistent with previous reports that demon-strated accumulation of long chain SM species, especially C16:0 and C18:0, in the liver, adipose tissue, or plasma following HFD or saturated fatty acid treatment [54,68,69]. These responses may indicate that with chronic HFD exposure, compensatory processes become activated to promote cell proliferation, dif-ferentiation, and liver regeneration [67]. On the other hand, reduced SMs (46.5%) could be associated with increased sphingomyelin hydrolysis via the sphingomyelinase pathway, yielding ceramides. Previ-ous studies reported that HFD mediated ceramide accumulation in liver was accompanied with increased sphingomyelinase activity [7,69]. In our mouse model, HFD prominently increased SM expression, which may have been hepatoprotective, whereas both reduced SMs and increased ceramides could exac-erbate liver injury.
Recent studies have reported that Glycosphingolipids (GSLs), in particular gangliosides such as GM3, participate in the pathological conditions of insulin resistance and hepatic steatosis. GSLs serve as regulators of transmembrane signaling to modulate cell proliferation, differentiation, and development. Insulin resistance induced by Tumor Necrosis Factor-Alpha (TNF-a) in adipocytes was accompanied by increased GM3 ganglioside [66]. Furthermore, genetic obesity models produced higher levels of GM3 synthase mRNA in adipose tissues relative to their lean counterparts indicating that GM3 may participate in the pathological conditions of insulin resistance in type 2 diabetes [66]. Mechanistic studies showed that certain gangliosides inhibit insulin receptor tyrosine kinase activity leading to inhibition of insulin-dependent cell growth and differentiation [67]. On the other hand, inhibition of GSL synthesis ameliorat-ed hepatic steatosis in obese mice by inhibiting fatty acid and TG synthesis and increasing ß-oxidation pathways [68]. Although our lipidomics analysis detected only two GSLs, one of them significantly was increased by HFD feeding suggesting a potential role in HFD-mediated hepatic insulin resistance and ste-atosis. This elevated GSL may have contributed to liver pathology by increasing lipotoxicity and reducing insulin sensitivity through inhibition of insulin receptor phosphorylation.
This study further validates the existing evidence that accumulation of TGs, DGs, and free cho-lesterol is considered to be the hallmark of NAFLD [8]. Our findings support this concept by detecting increased hepatic expression of TG species (55.6%) in HFD livers relative to controls. Liver fat accumulation occurs when the rate of hepatic triglyceride synthesis (hepatic fatty acid uptake and esterification into TG as well as de novo TG synthesis) exceeds the rate of triglyceride metabolism (fatty oxidation and TG export as Very Low Density Lipoproteins (VLDL)). Numerous studies conducted on vari-ous rodent models of obesity and obese humans have identified a correlation between hepatic lipid con-tent and the development of insulin resistance in liver. However, due to our focused lipidomics analysis with a mass range of 600-1200 Da, unlike TG, fewer DG species, were detected in the mouse livers. Our findings indicate that HFD resulted in a significant reduction in hepatic expression of DG(36:3); however, this may not represent the behavior of the entire DG profile. Further analysis to understand HFD mediated alterations on hepatic DG expression focused on a lower mass range (400-600 Da) awaits investigation.
In conclusion, this study demonstrates major abnormalities in hepatic phospholipid, sphingolipid, and glycerolipid expression in an experimental model of HFD induced NAFLD. A strength of the present study is the relative quantitative measurement of 17 lipid subclasses and 392 individual lipid ions in tan-dem. Our findings provide new insights into the lipid status of HFD mediated hepatic steatosis that could help clarify the molecular pathophysiology of NAFLD. The altered expression of hepatic phospholipids is likely to contribute to impaired membrane integrity and permeability leading to the progression of NAFLD, while altered sphingolipids may promote insulin resistance, inflammation, and liver injury. Further mechanistic studies of hepatic gene expression is influenced by phospholipid and sphingolipid me-tabolism would be necessary to explore potential mechanisms of action. Such studies may determine the therapeutic effects of either abstinence or pharmacologic treatment as a means to restore hepatic lipid ho-meostasis.
References
- Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Alimentary pharmacology & therapeutics. 2011; 34: 274-285.
- Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Asso-ciation. Hepatology (Baltimore, Md). 2012; 55: 2005-2023.
- Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y, et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology (Baltimore, Md). 2016; 64: 1577-1586.
- Shetty A, Syn WK. Current tretment options for nonalcoholic fattliver disease. Current opinion in gastroenterology. 2019; 35: 168-176.
- Sharma M, Mitnala S, Vishnubhotla RK, Mukherjee R, Reddy DN, Rao PN. The Riddle of Nonalcoholic Fatty Liver Disease: Progression from Nonalcoholic Fatty Liver to Nonalcoholic Steatohepatitis. Journal of clinical and experimental hepatology. 2015; 5: 147-158.
- Masarone M, Rosato V, Dallio M, Gravina AG, Aglitti A, et al. Role of Oxidative Stress in Patho-physiology of Nonalcoholic Fatty Liver Disease. Oxidative medicine and cellular longevity. 2018; 9547613: 1-14.
- Longato L, Tong M, Wands JR, de la Monte SM. High fat diet induced hepatic steatosis and insulin resistance: Role of dysregulated ceramide metabolism. Hepatology research: the official journal of the Japan Society of Hepatology. 2012; 42: 412-27.
- Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, et al. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. The Journal of clinical investigation. 2005; 115: 1343-1351.
- Ipsen DH, Lykkesfeldt J, Tveden-Nyborg P. Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cellular and molecular life sciences: CMLS. 2018; 75: 3313-3327.
- Lewis GF, Carpentier A, Adeli K, Giacca A. Disordered fat storage and mobilization in the pathogen-esis of insulin resistance and type 2 diabetes. Endocrine reviews. 2002; 23: 201-229.
- Cusi K. Role of insulin resistance and lipotoxicity in non-alcoholic steatohepatitis. Clinics in liver disease. 2009; 13: 545-563.
- Postic C, Girard J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin re-sistance: lessons from genetically engineered mice. The Journal of clinical investigation. 2008; 118: 829-838.
- Lambert JE, Ramos-Roman MA, Browning JD, Parks EJ. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology. 2014; 146: 726-735.
- Yamaguchi K, Yang L, McCall S, Huang J, Yu XX, Pandey SK, et al. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohep-atitis. Hepatology (Baltimore, Md). 2007; 45: 1366-1374.
- McClain CJ, Barve S, Deaciuc I. Good fat/bad fat. Hepatology (Baltimore, Md). 2007; 45: 1343-1346.
- Leamy AK, Egnatchik RA, Young JD. Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Progress in lipid research. 2013; 52: 165-174.
- Alkhouri N, Dixon LJ, Feldstein AE. Lipotoxicity in nonalcoholic fatty liver disease: not all lipids are created equal. Expert review of gastroenterology & hepatology. 2009; 3: 445-451.
- Puri P, Baillie RA, Wiest MM, Mirshahi F, Choudhury J, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology (Baltimore, Md). 2007; 46: 1081-1090.
- Pagadala M, Kasumov T, McCullough AJ, Zein NN, Kirwan JP. Role of ceramides in nonalcoholic fatty liver disease. Trends in endocrinology and metabolism: TEM. 2012; 23: 365-371.
- Nikolova-Karakashian M. Alcoholic and non-alcoholic fatty liver disease: Focus on ceramide. Ad-vances in biological regulation. 2018; 70: 40-50.
- Gorden DL, Ivanova PT, Myers DS, McIntyre JO, VanSaun MN, et al. Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; compari-son to a murine model. PloS one. 2011; 6: e22775.
- Malhi H, Gores GJ. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Semi-nars in liver disease. 2008; 28: 360-369.
- Tiwari-Heckler S, Gan-Schreier H, Stremmel W, Chamulitrat W, Pathil A. Circulating Phospholipid Patterns in NAFLD Patients Associated with a Combination of Metabolic Risk Factors. Nutrients. 2018; 10: 649.
- Li Z, Agellon LB, Allen TM, Umeda M, Jewell L, Mason A, et al. The ratio of phosphatidylcholine to phospha-tidylethanolamine influences membrane integrity and steatohepatitis. Cell metabolism. 2006; 3: 321-331.
- van der Veen JN, Kennelly JP, Wan S, Vance JE, Vance DE, et al. The critical role of phosphatidyl-choline and phosphatidylethanolamine metabolism in health and disease. Biochimica et biophysica acta Biomembranes. 2017; 1859: 1558-1572.
- Lydic TA, Goo YH. Lipidomics unveils the complexity of the lipidome in metabolic diseases. Clini-cal and translational medicine. 2018; 7: 4.
- Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annual re-view of biochemistry. 2011; 80: 301-325.
- Zemski Berry KA, Murphy RC, Kosmider B, Mason RJ. Lipidomic characterization and localization of phospholipids in the human lung. Journal of lipid research. 2017; 58: 926-933.
- Yalcin EB, de la Monte SM. Review of matrix-assisted laser desorption ionization-imaging mass spectrometry for lipid biochemical histopathology. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2015; 63: 762-771.
- Deininger SO, Cornett DS, Paape R, Becker M, Pineau C, et al. Normalization in MALDI-TOF imag-ing datasets of proteins: practical considerations. Analytical and bioanalytical chemistry. 2011; 401: 167-181.
- Gode D, Volmer DA. Lipid imaging by mass spectrometry - a review. Analyst. 2013; 138: 1289-1315.
- Soltwisch J, Kettling H, Vens-Cappell S, Wiegelmann M, Muthing J, Dreiswerd K. Mass spectrometry imag-ing with laser-induced postionization. Science (New York, NY). 2015; 348: 211-215.
- Cater CL, Jones JW, Farese AM, MacVittie TJ, Kane MA. Inflation-Fixation Method for Lipidomic Mapping of Lung Biopsies by Matrix Assisted Laser Desorption/Ionization-Mass Spectrometry Imag-ing. Analytical chemistry. 2016; 88: 4788-4794.
- Park ES, Lee JH, Hong JH, Park YK, Lee JW, Kim KP, et al. Phosphatidylcholine alteration identified using MALDI imaging MS in HBV-infected mouse livers and virus-mediated regeneration defects. PloS one. 2014; 9: e103955.
- Dreisewerd K, Lemaire R, Pohlentz G, Salzet M, Wisztorski M, Berkenkamp S, et al. Molecular profiling of native and matrix-coated tissue slices from rat brain by infrared and ultraviolet laser desorption/ionization orthogonal timeof- flight mass spectrometry. Analytical chemistry. 2007; 79: 2463-2471.
- Miyamura N, Nakamura T, Goto-Inoue N, Zaima N, Hayasaka T, Yamasaki T, et al. Imaging mass spectrometry reveals characteristic changes in triglyceride and phospholipid species in regenerating mouse liver. Biochemical and biophysical research communications. 2011; 408: 120-125.
- Ramanadham S, Hsu FF, Bohrer A, Nowatzke W, Ma Z, Turk J. Electrospray ionization mass spectro-metric analyses of phospholipids from rat and human pancreatic islets and subcellular membranes: comparison to other tissues and implications for membrane fusion in insulin exocytosis. Biochemis-try. 1998; 37: 4553-4567.
- Tyurin VA, Tyurina YY, Feng W, Mnuskin A, Jiang J, Tang M, et al. Massspectrometric characterization of phospholipids and their primary peroxidation products in rat cortical neurons during staurosporine-induced apoptosis. Journal of neurochemistry. 2008; 107: 1614-1633.
- Petkovic M, Schiller J, Muller M, Benard S, Reichl S, Arnold K, et al. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phos-phatidylcholine prevents the detection of further species. Analytical biochemistry. 2001; 289: 202-216.
- Afshinnia F, Rajendiran TM, Soni T, Byun J, Wernisch S, et al. Impaired beta- Oxidation and Altered Complex Lipid Fatty Acid Partitioning with Advancing CKD. Journal of the American Society of Nephrology: JASN. 2018; 29: 295- 306.
- Rao S, Walters KB, Wilson L, Chen B, Bolisetty S, Graves D, et al. Early lipid changes in acute kidney injury using SWATH lipidomics coupled with MALDI tissue imaging. American journal of physiology Re-nal physiology. 2016; 310: F1136-F147.
- Jackson SN, Barbacci D, Egan T, Lewis EK, Schultz JA, Woods A. MALDIIon Mobility Mass Spectrom-etry of Lipids in Negative Ion Mode. Analytical methods: advancing methods and applications. 2014; 6: 5001-5007.
- Busik JV, Reid GE, Lydic TA. Global analysis of retina lipids by complementary precursor ion and neutral loss mode tandem mass spectrometry. Methods in molecular biology (Clifton, NJ). 2009; 579: 33-70.
- Hsu FF, Turk J. Characterization of phosphatidylinositol, phosphatidylinositol- 4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mech-anistic study. Journal of the American Society for Mass Spectrometry. 2000; 11: 986-999.
- Lee CY, Lesimple A, Larsen A, Mamer O, Genest J. ESI-MS quantitation of increased sphingomyelin in Niemann-Pick disease type B HDL. Journal of lipid research. 2005; 46: 1213-1228.
- Hidaka H, Hanyu N, Sugano M, Kawasaki K, Yamauchi K, et al. Analysis of human serum lipopro-tein lipid composition using MALDI-TOF mass spectrometry. Annals of clinical and laboratory sci-ence. 2007; 37: 213-221.
- Hall Z, Bond NJ, Ashmore T, Sanders F, Ament Z, Wang X, et al. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology (Baltimore, Md). 2017; 65: 1165-1180.
- Shanta SR, Zhou LH, Park YS, Kim YH, Kim Y, et al. Binary matrix for MALDI imaging mass spec-trometry of phospholipids in both ion modes. Anal Chem. 2011; 83: 1252-1259.
- Debois D, Bralet MP, Le Naour F, Brunelle A, Laprevote O. In situ lipidomic analysis of nonalcohol-ic fatty liver by cluster TOF-SIMS imaging. Analytical chemistry. 2009; 81: 2823-2831.
- Nishikawa K, Hashimoto M, Itoh Y, Hiroi S, Lwaya K. Detection of changes in the structure and distribution map of triacylglycerol in fatty liver model by MALDI-SpiralTOF. FEBS open bio. 2014; 4: 179-184.
- Lee DY, Platt V, Bowen B, Louie K, Canaria CA, MC Murray CT, et al. Resolving brain regions using nanostructure initiator mass spectrometry imaging of phospholipids. Integrative biology: quantitative biosciences from nano to macro. 2012; 4: 693-699.
- Yalcin EB, Nunez K, Cornett DS, de la Monte SM. Differential Lipid Profiles in Experimental Stea-tohepatitis: Role for Imaging Mass Spectrometry as a Diagnostic Aid. J Drug Alcohol Res. 2015; 4: 1-11.
- Fisher-Wellman KH, Ryan TE, Smith CD, Gilliam LA, Lin CT, et al. A Direct Comparison of Meta-bolic Responses to High-Fat Diet in C57BL/6J and C57BL/6NJ Mice. Diabetes. 2016; 65: 3249-3261.
- Eisinger K, Liebisch G, Schmitz G, Aslanidis C, Krautbauer S, Buechler C. Lipidomic analysis of serum from high fat diet induced obese mice. International journal of molecular sciences. 2014; 15: 2991-3002.
- Dowhan W, Bogdanov M. Functional roles of lipids in membranes. Vance DE, Vance JE, editors. Amsterdam: Elsevier Science; 2002; 1-35.
- Anjani K, Lhomme M, Sokolovska N, Poitou C, Aron-Wisnewsky J, Bouillot JL, et al. Circulating phospholipid profiling identifies portal contribution to NASH signature in obesity. Journal of Hepatology. 2015; 62: 905-912.
- Matsuda S, Kobayashi M, Kitagishi Y. Roles for PI3K/AKT/PTEN Pathway in Cell Signaling of Nonalcoholic Fatty Liver Disease. ISRN endocrinology. 2013; 2013: 472432.
- Nagata S. Apoptosis and autoimmune diseases. Annals of the New York Academy of Sciences. 2010; 1209: 10-16.
- Ma DW, Arendt BM, Hillyer LM, Fung SK, McGilvray I, Allard J. Plasma phospholipids and fatty acid composition differ between liver biopsy-proven nonalcoholic fatty liver disease and healthy subjects. Nutrition & diabetes. 2016; 6: e220.
- Huang BX, Akbar M, Kevala K, Kim HY. Phosphatidylserine is a critical modulator for Akt activa-tion. J cell biol. 2011; 192: 979-992.
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiological reviews. 2013; 93: 1019-1137.
- Spooner MH, Jump DB. Omega-3 fatty acids and nonalcoholic fatty liver disease in adults and chil-dren: where do we stand? Current opinion in clinical nutrition and metabolic care. 2019; 22: 103-110.
- Jump DB, Lytle KA, Depner CM, Tripathy S. Omega-3 polyunsaturated fatty acids as a treatment strategy for nonalcoholic fatty liver disease. Pharmacology & therapeutics. 2018; 181: 108-125.
- Jump DB, Botolin D, Wang Y, Xu J, Demeure O, Christian B. Docosahexaenoic Acid (DHA) and hepatic gene transcription. Chemistry and physics of lipids. 2008; 153: 3-13.
- Balsinde J, Winstead MV, Dennis EA. Phospholipase A(2) regulation of arachidonic acid mobiliza-tion. FEBS letters. 2002; 531: 2-6.
- Tagami S, Inokuchi Ji J, Kabayama K, Yoshimura H, Kitamura F, Uemura S, et al. Ganglioside GM3 partici-pates in the pathological conditions of insulin resistance. The Journal of biological chemistry. 2002; 277: 3085-3092.
- Nojiri H, Stroud M, Hakomori S. A specific type of ganglioside as a modulator of insulin-dependent cell growth and insulin receptor tyrosine kinase activity. Possible association of ganglioside-induced inhibition of insulin receptor function and monocytic differentiation induction in HL-60 cells. The Journal of biological chemistry. 1991; 266: 4531-4537.
- Zhao H, Przybylska M, Wu IH, Zhang J, Maniatis P, Pacheco J, et al. Inhibiting glycosphingolipid synthesis ameliorates hepatic steatosis in obese mice. Hepatology (Baltimore, Md). 2009; 50: 85-93.
